This adds a dosing event to the event table. This is provided for
piping syntax through magrittr. It can also be accessed by eventTable$add.dosing(...)
add.dosing(
eventTable,
dose,
nbr.doses = 1L,
dosing.interval = 24,
dosing.to = 1L,
rate = NULL,
amount.units = NA_character_,
start.time = 0,
do.sampling = FALSE,
time.units = NA_character_,
...
)Arguments
| eventTable | eventTable object; When accessed from object it would be |
|---|---|
| dose | numeric scalar, dose amount in |
| nbr.doses | integer, number of doses; |
| dosing.interval | required numeric scalar, time between doses
in |
| dosing.to | integer, compartment the dose goes into (first compartment by default); |
| rate | for infusions, the rate of infusion (default is
|
| amount.units | optional string indicating the dosing units.
Defaults to |
| start.time | required dosing start time; |
| do.sampling | logical, should observation sampling records be
added at the dosing times? Defaults to |
| time.units | optional string indicating the time units.
Defaults to |
| ... | Other parameters passed to |
Value
eventTable with updated dosing (note the event table will be updated anyway)
References
Wang W, Hallow K, James D (2015). "A Tutorial on RxODE: Simulating Differential Equation Pharmacometric Models in R." CPT: Pharmacometrics \& Systems Pharmacology, 5(1), 3-10. ISSN 2163-8306, <URL: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4728294/>.
See also
eventTable, add.sampling,
add.dosing, et,
etRep, etRbind,
RxODE
Author
Matthew L. Fidler
Matthew L Fidler, Wenping Wang
Examples
# \donttest{
library(RxODE)
library(units)
#> udunits database from /usr/share/xml/udunits/udunits2.xml
## Model from RxODE tutorial
mod1 <-RxODE({
KA=2.94E-01;
CL=1.86E+01;
V2=4.02E+01;
Q=1.05E+01;
V3=2.97E+02;
Kin=1;
Kout=1;
EC50=200;
C2 = centr/V2;
C3 = peri/V3;
d/dt(depot) =-KA*depot;
d/dt(centr) = KA*depot - CL*C2 - Q*C2 + Q*C3;
d/dt(peri) = Q*C2 - Q*C3;
d/dt(eff) = Kin - Kout*(1-C2/(EC50+C2))*eff;
});
#>
## These are making the more complex regimens of the RxODE tutorial
## bid for 5 days
bid <- et(timeUnits="hr") %>%
et(amt=10000,ii=12,until=set_units(5, "days"))
## qd for 5 days
qd <- et(timeUnits="hr") %>%
et(amt=20000,ii=24,until=set_units(5, "days"))
## bid for 5 days followed by qd for 5 days
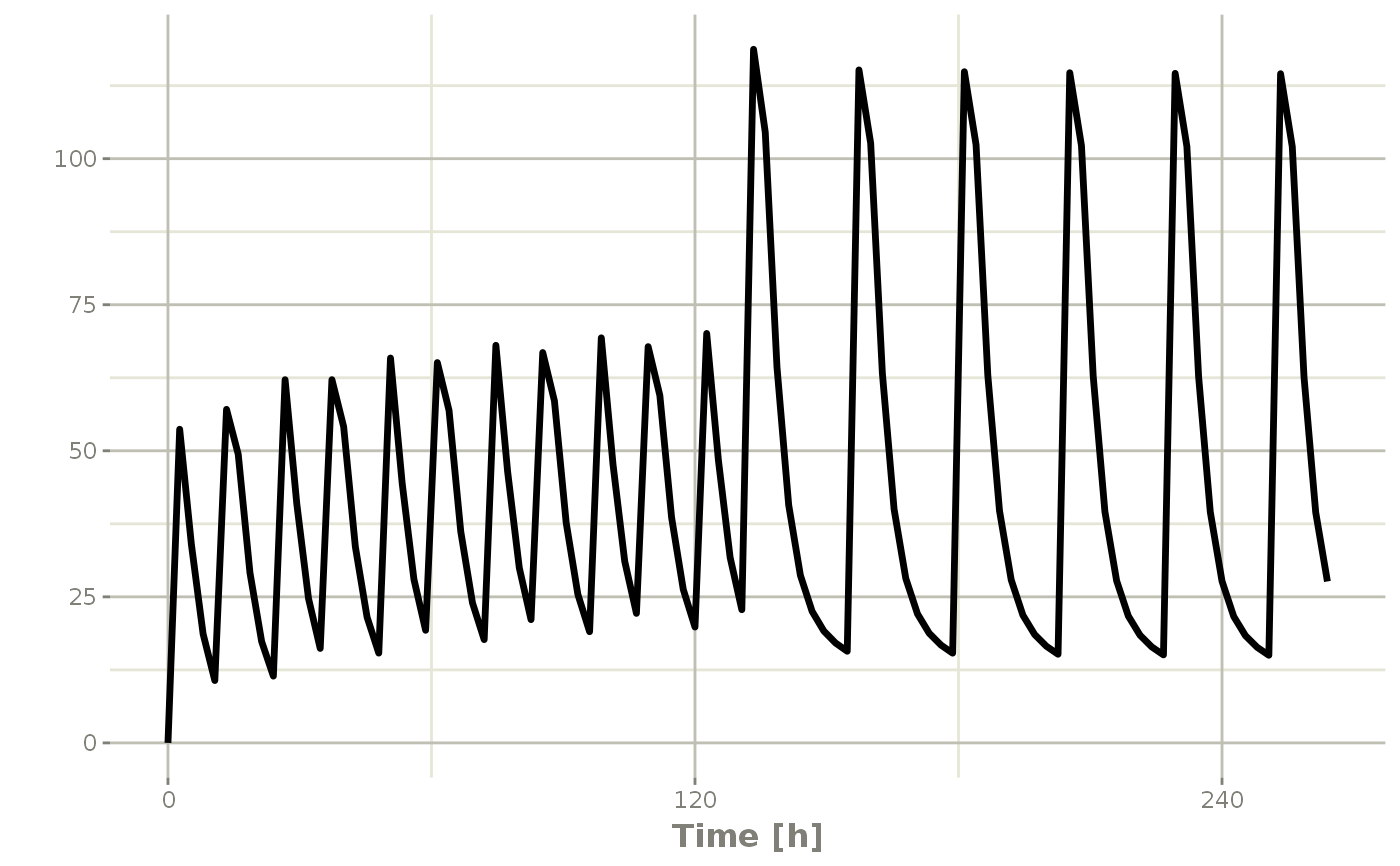
et <- seq(bid,qd) %>% et(seq(0,11*24,length.out=100));
bidQd <- rxSolve(mod1, et)
plot(bidQd, C2)
 ## Now Infusion for 5 days followed by oral for 5 days
## note you can dose to a named compartment instead of using the compartment number
infusion <- et(timeUnits = "hr") %>%
et(amt=10000, rate=5000, ii=24, until=set_units(5, "days"), cmt="centr")
qd <- et(timeUnits = "hr") %>% et(amt=10000, ii=24, until=set_units(5, "days"), cmt="depot")
et <- seq(infusion,qd)
infusionQd <- rxSolve(mod1, et)
plot(infusionQd, C2)
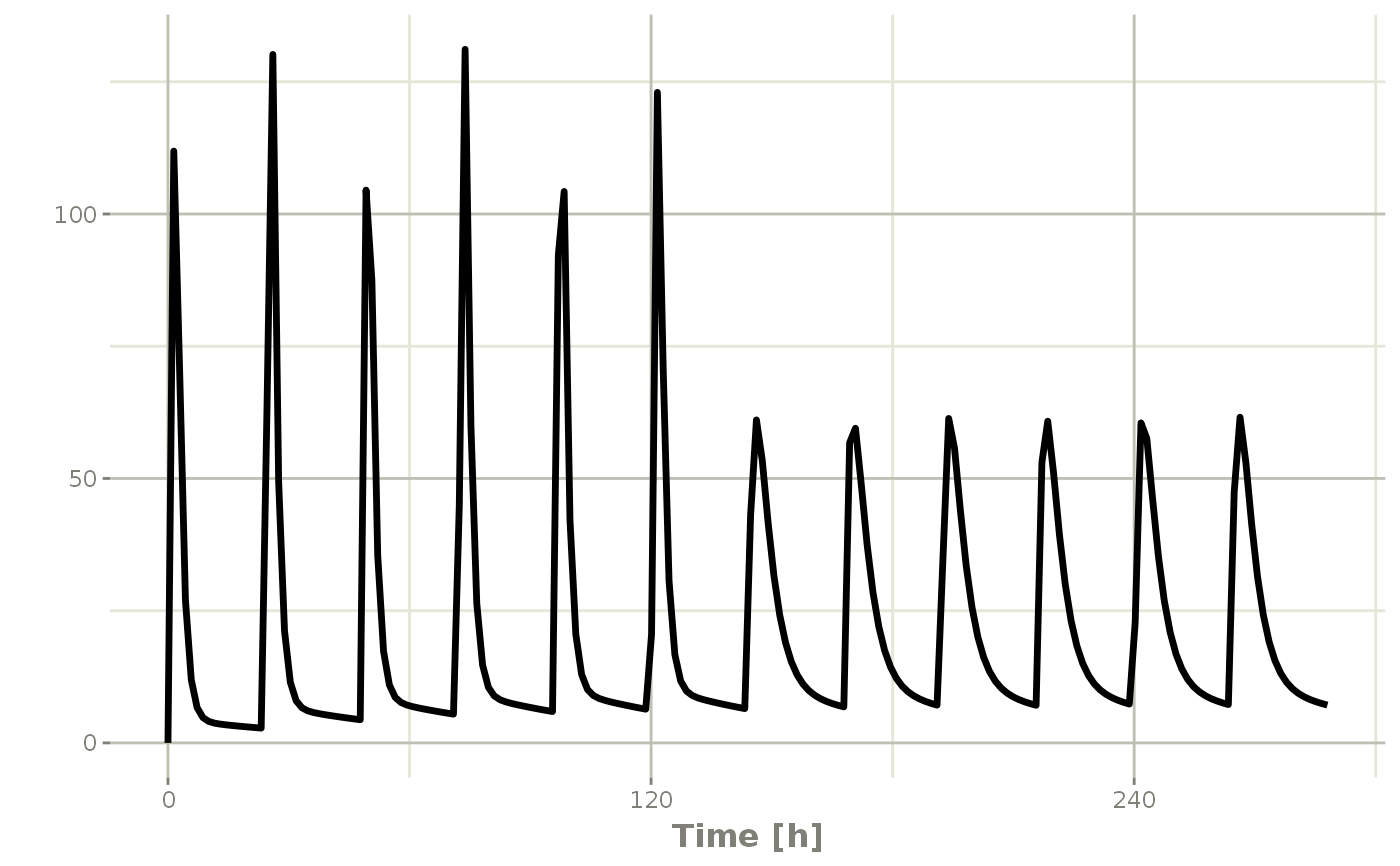
## Now Infusion for 5 days followed by oral for 5 days
## note you can dose to a named compartment instead of using the compartment number
infusion <- et(timeUnits = "hr") %>%
et(amt=10000, rate=5000, ii=24, until=set_units(5, "days"), cmt="centr")
qd <- et(timeUnits = "hr") %>% et(amt=10000, ii=24, until=set_units(5, "days"), cmt="depot")
et <- seq(infusion,qd)
infusionQd <- rxSolve(mod1, et)
plot(infusionQd, C2)
 ## 2wk-on, 1wk-off
qd <- et(timeUnits = "hr") %>% et(amt=10000, ii=24, until=set_units(2, "weeks"), cmt="depot")
et <- seq(qd, set_units(1,"weeks"), qd) %>%
add.sampling(set_units(seq(0, 5.5,by=0.005),weeks))
wkOnOff <- rxSolve(mod1, et)
plot(wkOnOff, C2)
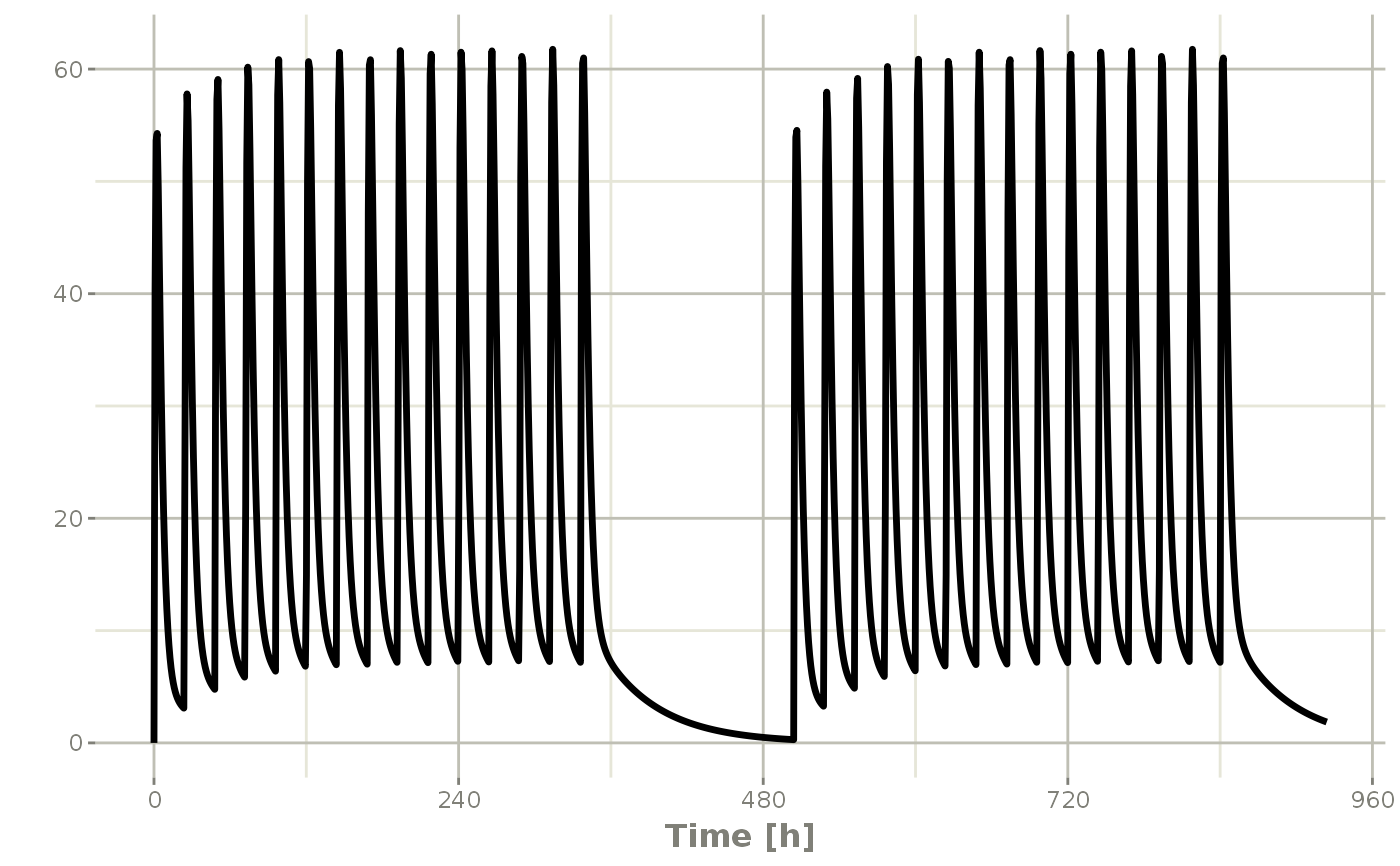
## 2wk-on, 1wk-off
qd <- et(timeUnits = "hr") %>% et(amt=10000, ii=24, until=set_units(2, "weeks"), cmt="depot")
et <- seq(qd, set_units(1,"weeks"), qd) %>%
add.sampling(set_units(seq(0, 5.5,by=0.005),weeks))
wkOnOff <- rxSolve(mod1, et)
plot(wkOnOff, C2)
 ## You can also repeat the cycle easily with the rep function
qd <-et(timeUnits = "hr") %>% et(amt=10000, ii=24, until=set_units(2, "weeks"), cmt="depot")
et <- etRep(qd, times=4, wait=set_units(1,"weeks")) %>%
add.sampling(set_units(seq(0, 12.5,by=0.005),weeks))
repCycle4 <- rxSolve(mod1, et)
plot(repCycle4, C2)
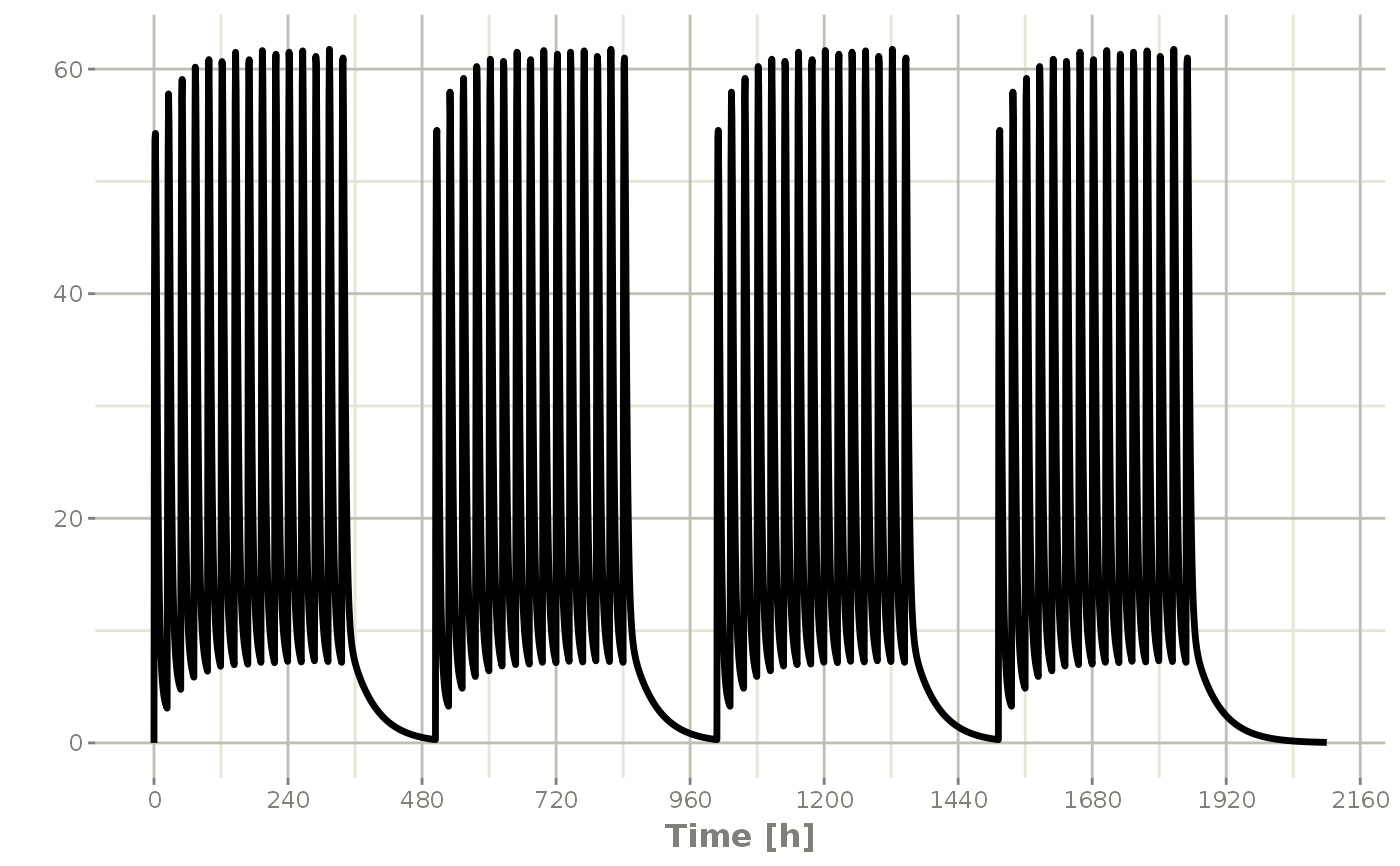
## You can also repeat the cycle easily with the rep function
qd <-et(timeUnits = "hr") %>% et(amt=10000, ii=24, until=set_units(2, "weeks"), cmt="depot")
et <- etRep(qd, times=4, wait=set_units(1,"weeks")) %>%
add.sampling(set_units(seq(0, 12.5,by=0.005),weeks))
repCycle4 <- rxSolve(mod1, et)
plot(repCycle4, C2)
 # }
# }