Using RxODE data frames
Creating an interactive data frame
RxODE supports returning a solved object that is a modified data-frame. This is done by the predict(), solve(), or rxSolve() methods.
library(RxODE)
library(units)
## Setup example model
mod1 <-RxODE({
C2 = centr/V2;
C3 = peri/V3;
d/dt(depot) =-KA*depot;
d/dt(centr) = KA*depot - CL*C2 - Q*C2 + Q*C3;
d/dt(peri) = Q*C2 - Q*C3;
d/dt(eff) = Kin - Kout*(1-C2/(EC50+C2))*eff;
})
## Seup parameters and initial conditions
theta <-
c(KA=2.94E-01, CL=1.86E+01, V2=4.02E+01, # central
Q=1.05E+01, V3=2.97E+02, # peripheral
Kin=1, Kout=1, EC50=200) # effects
inits <- c(eff=1)
## Setup dosing event information
ev <- eventTable(amount.units="mg", time.units="hours") %>%
add.dosing(dose=10000, nbr.doses=10, dosing.interval=12) %>%
add.dosing(dose=20000, nbr.doses=5, start.time=120,
dosing.interval=24) %>%
add.sampling(0:240);
## Now solve
x <- predict(mod1,theta, ev, inits)
print(x)
#> ______________________________ Solved RxODE object _____________________________
#> -- Parameters ($params): -------------------------------------------------------
#> V2 V3 KA CL Q Kin Kout EC50
#> 40.200 297.000 0.294 18.600 10.500 1.000 1.000 200.000
#> -- Initial Conditions ($inits): ------------------------------------------------
#> depot centr peri eff
#> 0 0 0 1
#> -- First part of data (object): ------------------------------------------------
#> # A tibble: 241 x 7
#> time C2 C3 depot centr peri eff
#> [h] <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 0 0 10000 0 0 1
#> 2 1 44.4 0.920 7453. 1784. 273. 1.08
#> 3 2 54.9 2.67 5554. 2206. 794. 1.18
#> 4 3 51.9 4.46 4140. 2087. 1324. 1.23
#> 5 4 44.5 5.98 3085. 1789. 1776. 1.23
#> 6 5 36.5 7.18 2299. 1467. 2132. 1.21
#> # ... with 235 more rows
#> ________________________________________________________________________________or
x <- solve(mod1,theta, ev, inits)
print(x)
#> ______________________________ Solved RxODE object _____________________________
#> -- Parameters ($params): -------------------------------------------------------
#> V2 V3 KA CL Q Kin Kout EC50
#> 40.200 297.000 0.294 18.600 10.500 1.000 1.000 200.000
#> -- Initial Conditions ($inits): ------------------------------------------------
#> depot centr peri eff
#> 0 0 0 1
#> -- First part of data (object): ------------------------------------------------
#> # A tibble: 241 x 7
#> time C2 C3 depot centr peri eff
#> [h] <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 0 0 10000 0 0 1
#> 2 1 44.4 0.920 7453. 1784. 273. 1.08
#> 3 2 54.9 2.67 5554. 2206. 794. 1.18
#> 4 3 51.9 4.46 4140. 2087. 1324. 1.23
#> 5 4 44.5 5.98 3085. 1789. 1776. 1.23
#> 6 5 36.5 7.18 2299. 1467. 2132. 1.21
#> # ... with 235 more rows
#> ________________________________________________________________________________Or with mattigr
x <- mod1 %>% solve(theta, ev, inits)
print(x)
#> ______________________________ Solved RxODE object _____________________________
#> -- Parameters ($params): -------------------------------------------------------
#> V2 V3 KA CL Q Kin Kout EC50
#> 40.200 297.000 0.294 18.600 10.500 1.000 1.000 200.000
#> -- Initial Conditions ($inits): ------------------------------------------------
#> depot centr peri eff
#> 0 0 0 1
#> -- First part of data (object): ------------------------------------------------
#> # A tibble: 241 x 7
#> time C2 C3 depot centr peri eff
#> [h] <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 0 0 10000 0 0 1
#> 2 1 44.4 0.920 7453. 1784. 273. 1.08
#> 3 2 54.9 2.67 5554. 2206. 794. 1.18
#> 4 3 51.9 4.46 4140. 2087. 1324. 1.23
#> 5 4 44.5 5.98 3085. 1789. 1776. 1.23
#> 6 5 36.5 7.18 2299. 1467. 2132. 1.21
#> # ... with 235 more rows
#> ________________________________________________________________________________RxODE solved object properties
Using the solved object as a simple data frame
The solved object acts as a data.frame or tbl that can be filtered by dpylr. For example you could filter it easily.
library(dplyr)
## You can drop units for comparisons and filtering
x <- mod1 %>% solve(theta,ev,inits) %>%
drop_units %>% filter(time <= 3) %>% as.tbl
## or keep them and compare with the proper units.
x <- mod1 %>% solve(theta,ev,inits) %>%
filter(time <= set_units(3, hr)) %>% as.tbl
x
#> # A tibble: 4 x 7
#> time C2 C3 depot centr peri eff
#> [h] <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 0 0 10000 0 0 1
#> 2 1 44.4 0.920 7453. 1784. 273. 1.08
#> 3 2 54.9 2.67 5554. 2206. 794. 1.18
#> 4 3 51.9 4.46 4140. 2087. 1324. 1.23Updating the data-set interactively
However it isn’t just a simple data object. You can use the solved object to update parameters on the fly, or even change the sampling time.
First we need to recreate the original solved system:
x <- mod1 %>% solve(theta,ev,inits);
print(x)
#> ______________________________ Solved RxODE object _____________________________
#> -- Parameters ($params): -------------------------------------------------------
#> V2 V3 KA CL Q Kin Kout EC50
#> 40.200 297.000 0.294 18.600 10.500 1.000 1.000 200.000
#> -- Initial Conditions ($inits): ------------------------------------------------
#> depot centr peri eff
#> 0 0 0 1
#> -- First part of data (object): ------------------------------------------------
#> # A tibble: 241 x 7
#> time C2 C3 depot centr peri eff
#> [h] <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 0 0 10000 0 0 1
#> 2 1 44.4 0.920 7453. 1784. 273. 1.08
#> 3 2 54.9 2.67 5554. 2206. 794. 1.18
#> 4 3 51.9 4.46 4140. 2087. 1324. 1.23
#> 5 4 44.5 5.98 3085. 1789. 1776. 1.23
#> 6 5 36.5 7.18 2299. 1467. 2132. 1.21
#> # ... with 235 more rows
#> ________________________________________________________________________________Modifying initial conditions
To examine or change initial conditions, you can use the syntax cmt.0, cmt0, or cmt_0. In the case of the eff compartment defined by the model, this is:
x$eff0
#> [1] 1which shows the initial condition of the effect compartment. If you wished to change this initial condition to 2, this can be done easily by:
x$eff0 <- 2
print(x)
#> ______________________________ Solved RxODE object _____________________________
#> -- Parameters ($params): -------------------------------------------------------
#> V2 V3 KA CL Q Kin Kout EC50
#> 40.200 297.000 0.294 18.600 10.500 1.000 1.000 200.000
#> -- Initial Conditions ($inits): ------------------------------------------------
#> depot centr peri eff
#> 0 0 0 2
#> -- First part of data (object): ------------------------------------------------
#> # A tibble: 241 x 7
#> time C2 C3 depot centr peri eff
#> [h] <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 0 0 10000 0 0 2
#> 2 1 44.4 0.920 7453. 1784. 273. 1.50
#> 3 2 54.9 2.67 5554. 2206. 794. 1.37
#> 4 3 51.9 4.46 4140. 2087. 1324. 1.31
#> 5 4 44.5 5.98 3085. 1789. 1776. 1.27
#> 6 5 36.5 7.18 2299. 1467. 2132. 1.23
#> # ... with 235 more rows
#> ________________________________________________________________________________
plot(x)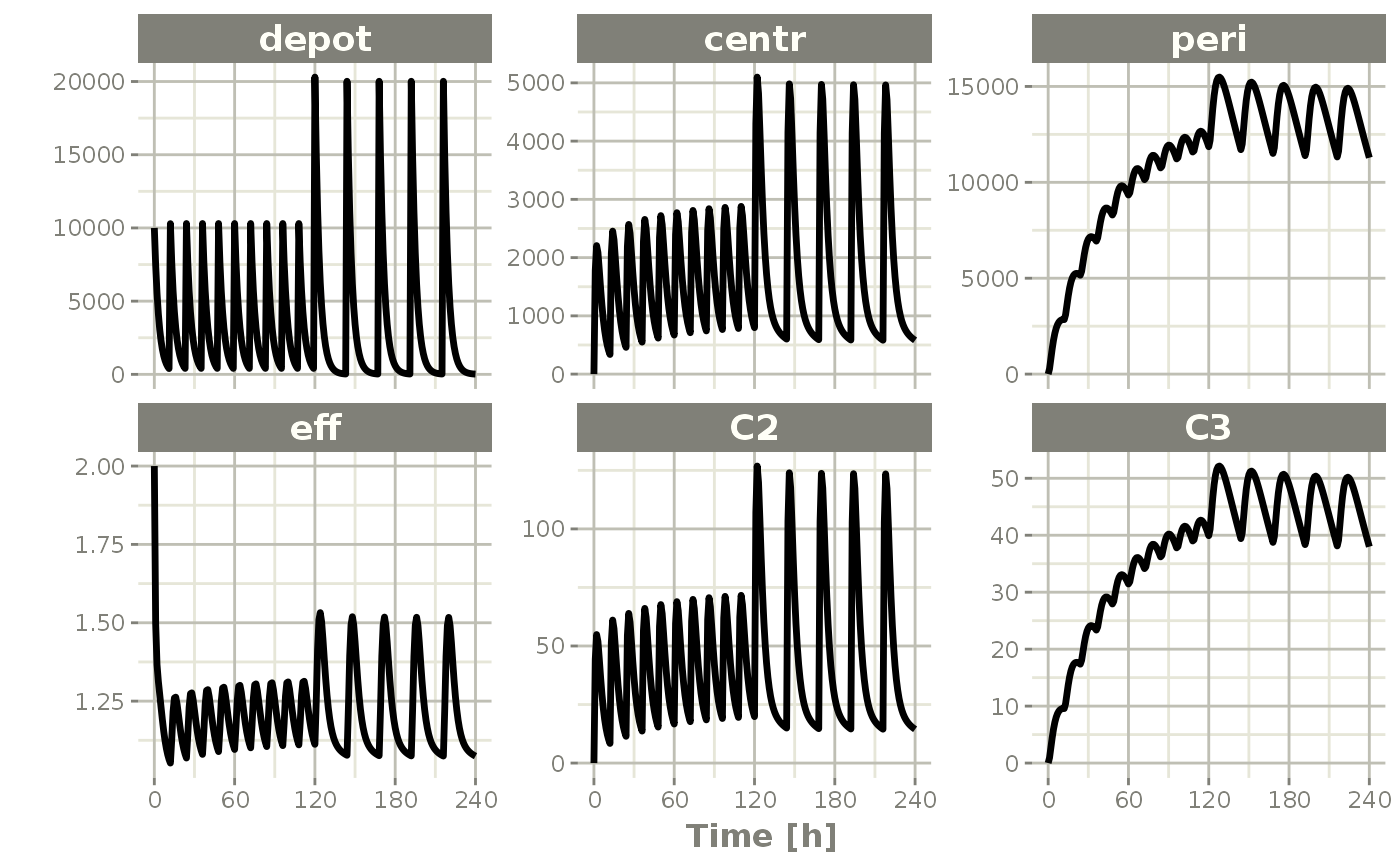
Modifying observation times for RxODE
Notice that the initial effect is now 2.
You can also change the sampling times easily by this method by changing t or time. For example:
x$t <- seq(0,5,length.out=20)
print(x)
#> ______________________________ Solved RxODE object _____________________________
#> -- Parameters ($params): -------------------------------------------------------
#> V2 V3 KA CL Q Kin Kout EC50
#> 40.200 297.000 0.294 18.600 10.500 1.000 1.000 200.000
#> -- Initial Conditions ($inits): ------------------------------------------------
#> depot centr peri eff
#> 0 0 0 2
#> -- First part of data (object): ------------------------------------------------
#> # A tibble: 20 x 7
#> time C2 C3 depot centr peri eff
#> [h] <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 0 0 10000 0 0 2
#> 2 0.263 16.8 0.0817 9255. 677. 24.3 1.79
#> 3 0.526 29.5 0.299 8566. 1187. 88.7 1.65
#> 4 0.789 38.9 0.615 7929. 1562. 183. 1.55
#> 5 1.05 45.5 1.00 7338. 1830. 298. 1.49
#> 6 1.32 50.1 1.44 6792. 2013. 427. 1.44
#> # ... with 14 more rows
#> ________________________________________________________________________________
plot(x)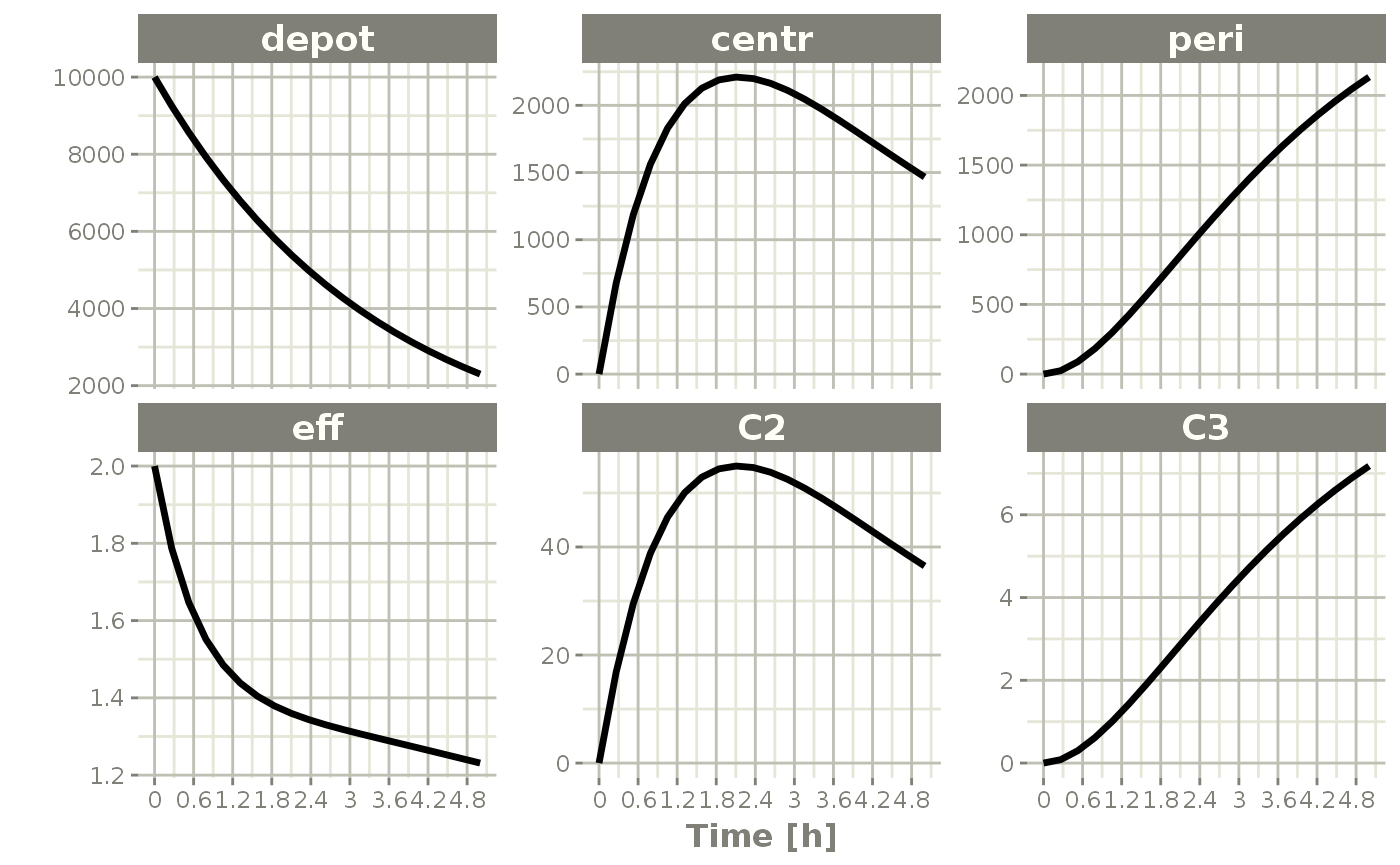
Modifying simulation parameters
You can also access or change parameters by the $ operator. For example, accessing KA can be done by:
x$KA
#> [1] 0.294And you may change it by assigning it to a new value.
x$KA <- 1
print(x)
#> ______________________________ Solved RxODE object _____________________________
#> -- Parameters ($params): -------------------------------------------------------
#> V2 V3 KA CL Q Kin Kout EC50
#> 40.2 297.0 1.0 18.6 10.5 1.0 1.0 200.0
#> -- Initial Conditions ($inits): ------------------------------------------------
#> depot centr peri eff
#> 0 0 0 2
#> -- First part of data (object): ------------------------------------------------
#> # A tibble: 20 x 7
#> time C2 C3 depot centr peri eff
#> [h] <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 0 0 10000 0 0 2
#> 2 0.263 52.2 0.261 7686. 2098. 77.6 1.82
#> 3 0.526 83.3 0.900 5908. 3348. 267. 1.74
#> 4 0.789 99.8 1.75 4541. 4010. 519. 1.69
#> 5 1.05 106. 2.69 3490. 4273. 800. 1.67
#> 6 1.32 106. 3.66 2683. 4272. 1086. 1.64
#> # ... with 14 more rows
#> ________________________________________________________________________________
plot(x)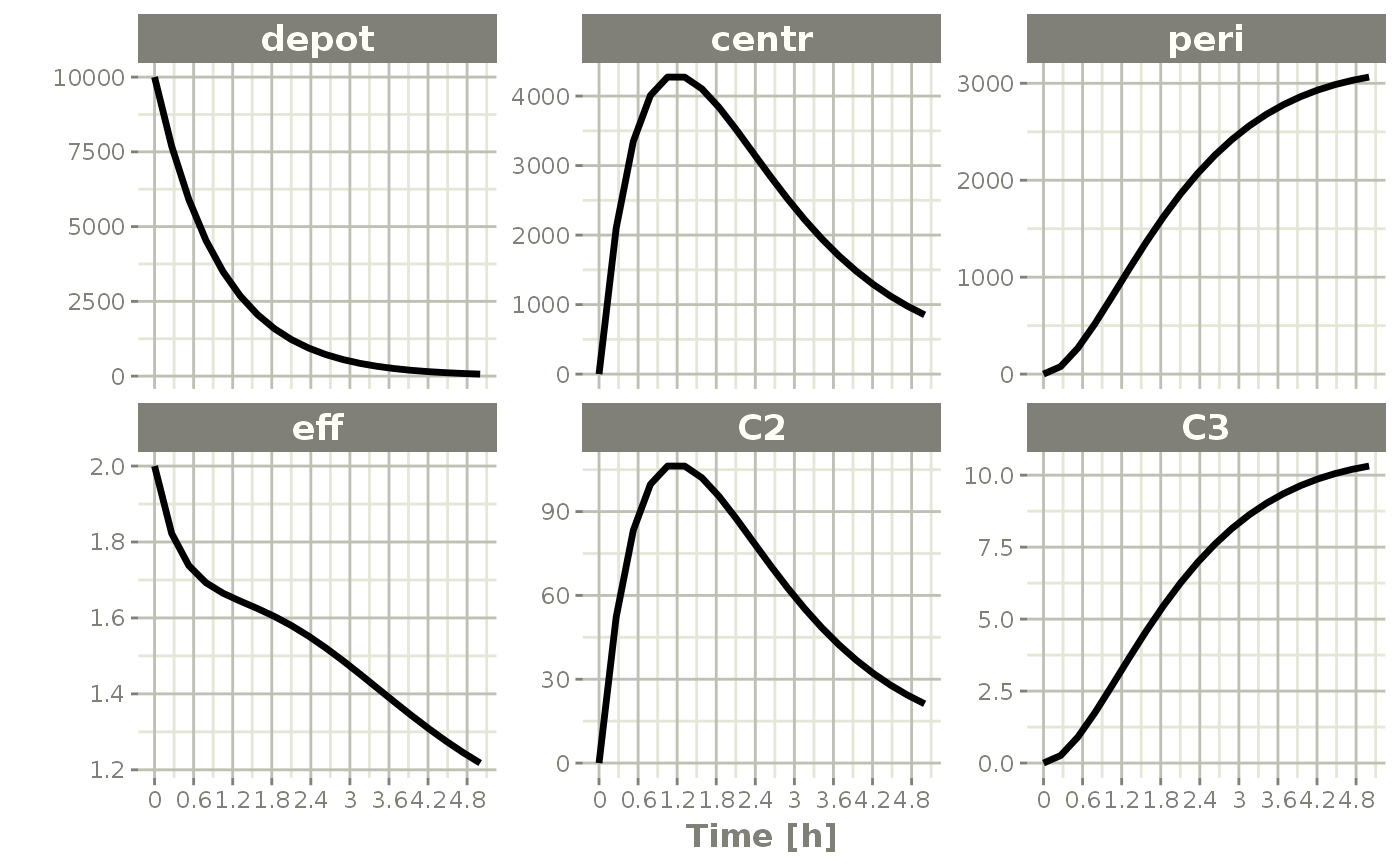
You can access/change all the parameters, initialization(s) or events with the $params, $inits, $events accessor syntax, similar to what is used above.
This syntax makes it easy to update and explore the effect of various parameters on the solved object.