wbc <- function() {
ini({
## Note that the UI can take expressions
## Also note that these initial estimates should be provided on the log-scale
log_CIRC0 <- log(7.21)
log_MTT <- log(124)
log_SLOPU <- log(28.9)
log_GAMMA <- log(0.239)
## Initial estimates should be high for SAEM ETAs
eta.CIRC0 ~ .1
eta.MTT ~ .03
eta.SLOPU ~ .2
## Also true for additive error (also ignored in SAEM)
prop.err <- 10
})
model({
CIRC0 = exp(log_CIRC0 + eta.CIRC0)
MTT = exp(log_MTT + eta.MTT)
SLOPU = exp(log_SLOPU + eta.SLOPU)
GAMMA = exp(log_GAMMA)
# PK parameters from input dataset
CL = CLI;
V1 = V1I;
V2 = V2I;
Q = 204;
CONC = A_centr/V1;
# PD parameters
NN = 3;
KTR = (NN + 1)/MTT;
EDRUG = 1 - SLOPU * CONC;
FDBK = (CIRC0 / A_circ)^GAMMA;
CIRC = A_circ;
A_prol(0) = CIRC0;
A_tr1(0) = CIRC0;
A_tr2(0) = CIRC0;
A_tr3(0) = CIRC0;
A_circ(0) = CIRC0;
d/dt(A_centr) = A_periph * Q/V2 - A_centr * (CL/V1 + Q/V1);
d/dt(A_periph) = A_centr * Q/V1 - A_periph * Q/V2;
d/dt(A_prol) = KTR * A_prol * EDRUG * FDBK - KTR * A_prol;
d/dt(A_tr1) = KTR * A_prol - KTR * A_tr1;
d/dt(A_tr2) = KTR * A_tr1 - KTR * A_tr2;
d/dt(A_tr3) = KTR * A_tr2 - KTR * A_tr3;
d/dt(A_circ) = KTR * A_tr3 - KTR * A_circ;
CIRC ~ prop(prop.err)
})
}Fit model using saem
d3 = read.csv("Simulated_WBC_pacl_ddmore_samePK_nlmixr.csv", na.string = ".")
fit.S <- nlmixr(wbc, d3, est="saem", list(print=0), table=list(cwres=TRUE, npde=TRUE))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "V2I" "CLI" "V1I"
#> [====|====|====|====|====|====|====|====|====|====] 0:00:04
xpdb <- xpose_data_nlmixr(fit.S)
plot(fit.S)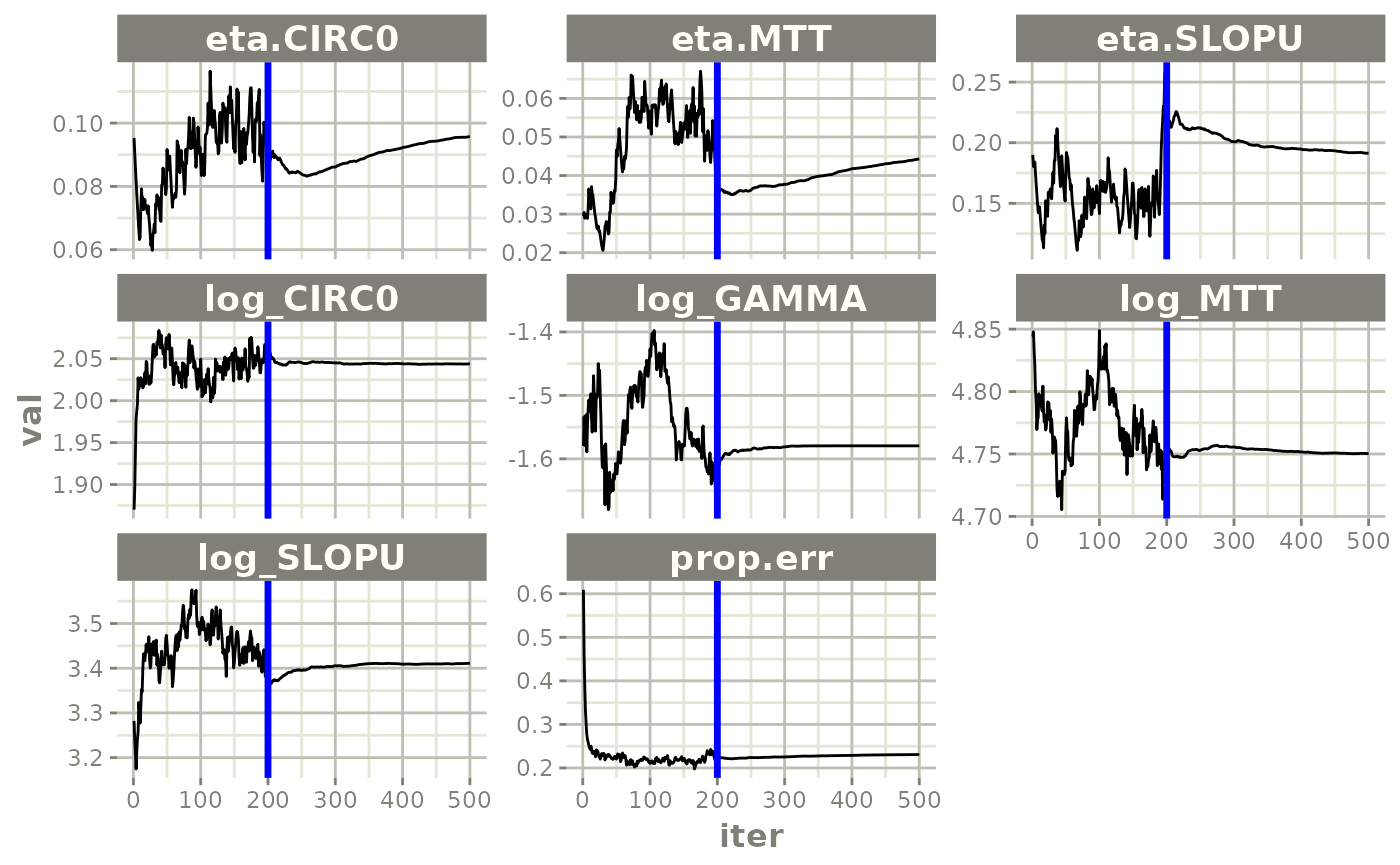
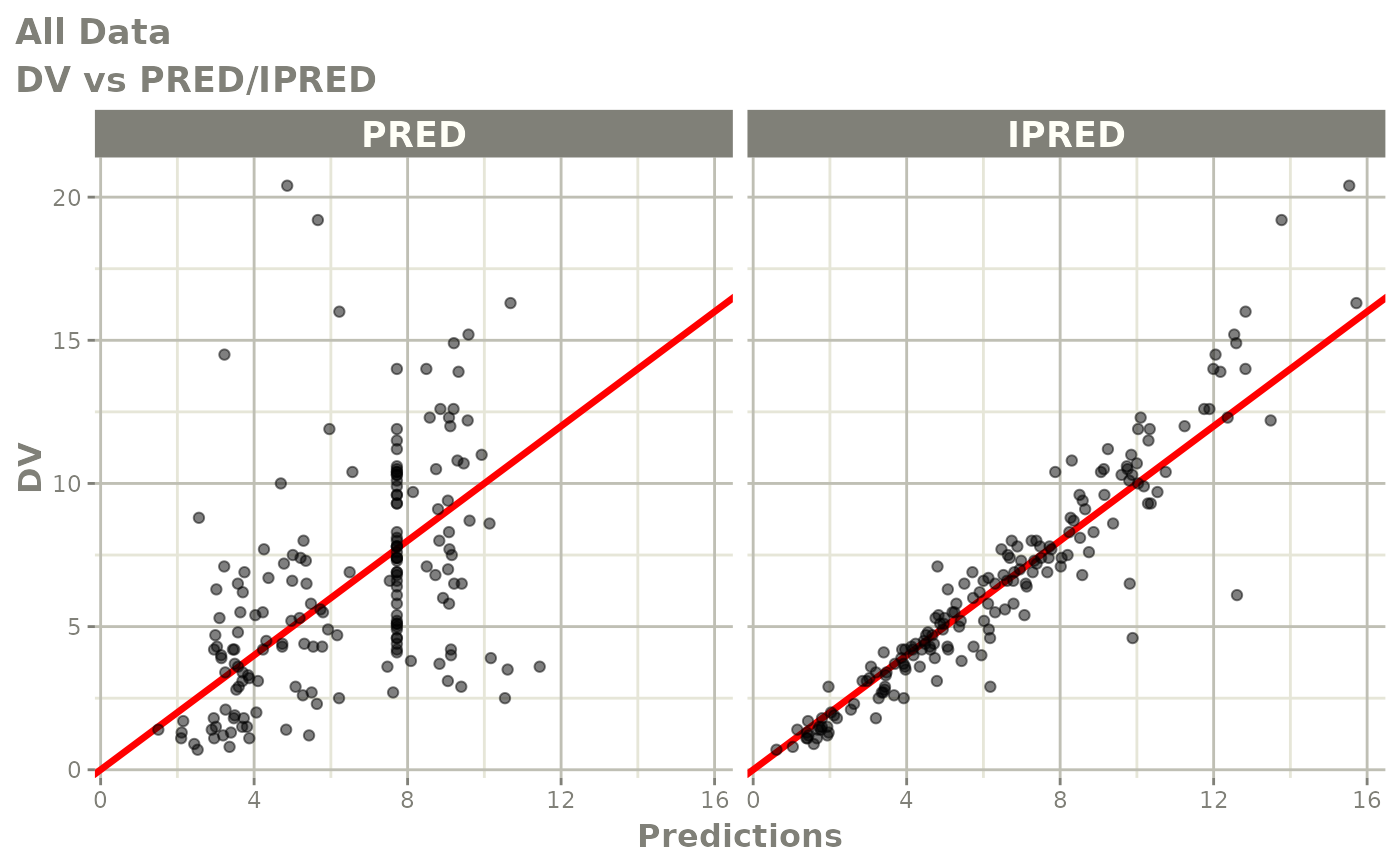
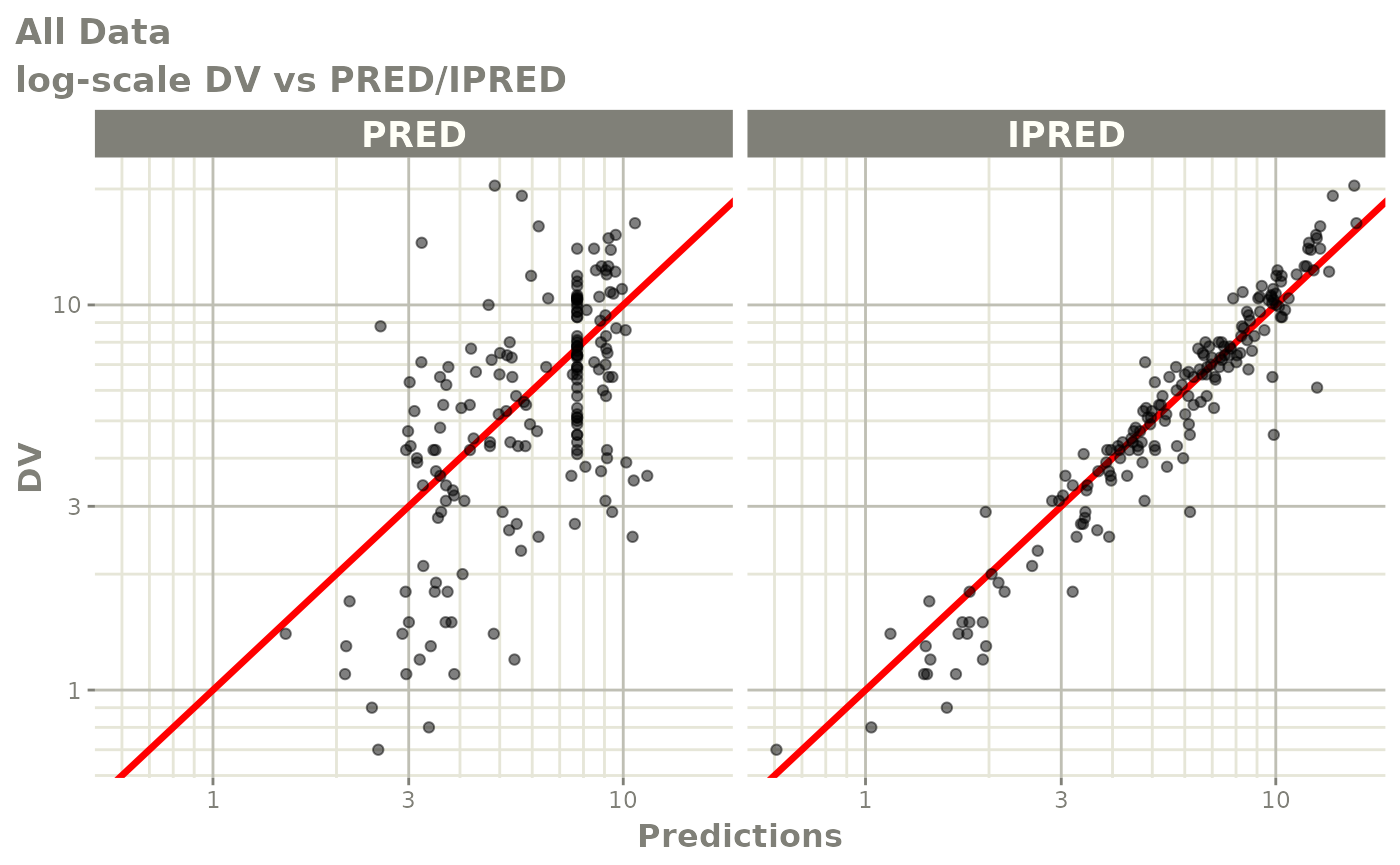
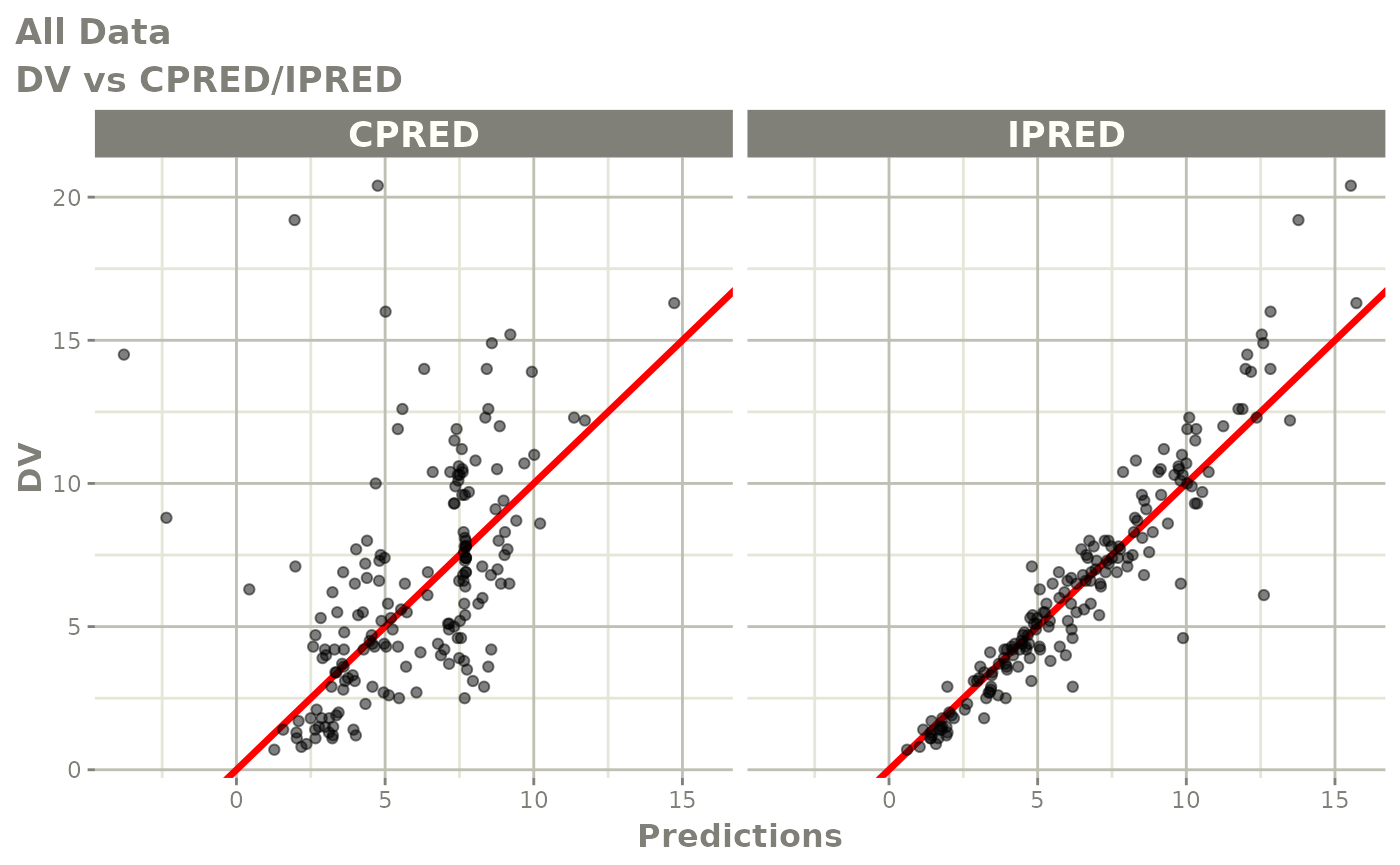
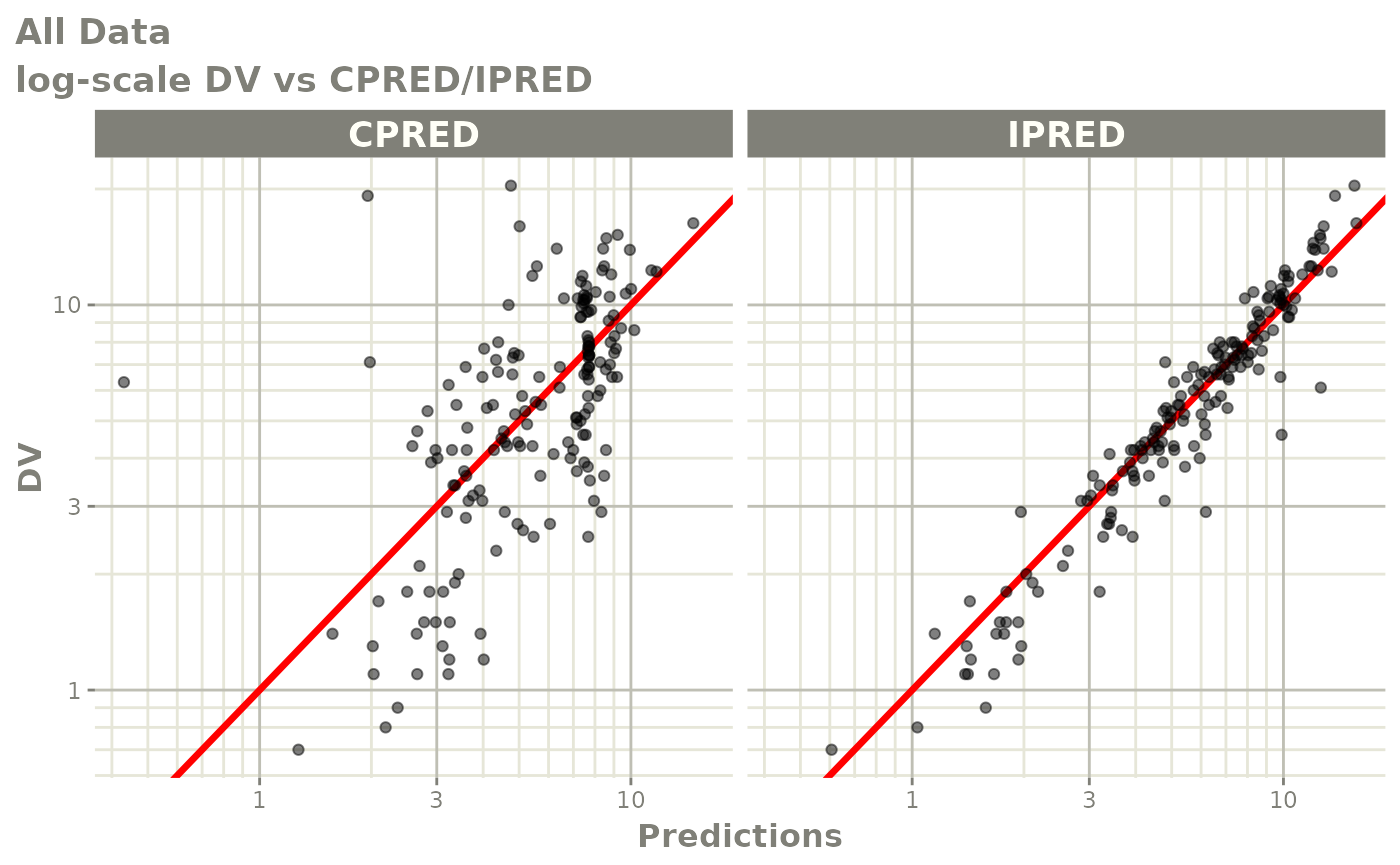
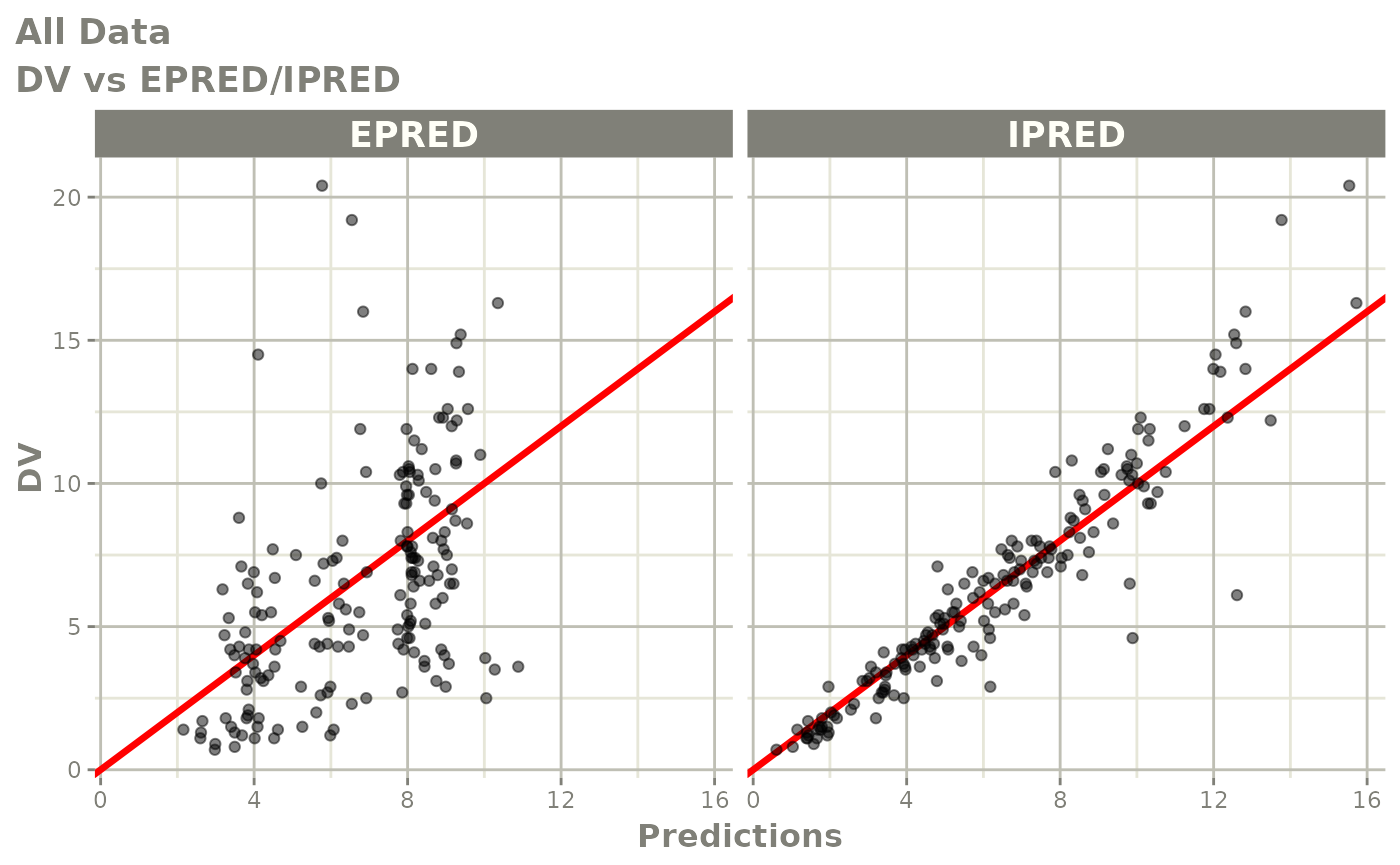
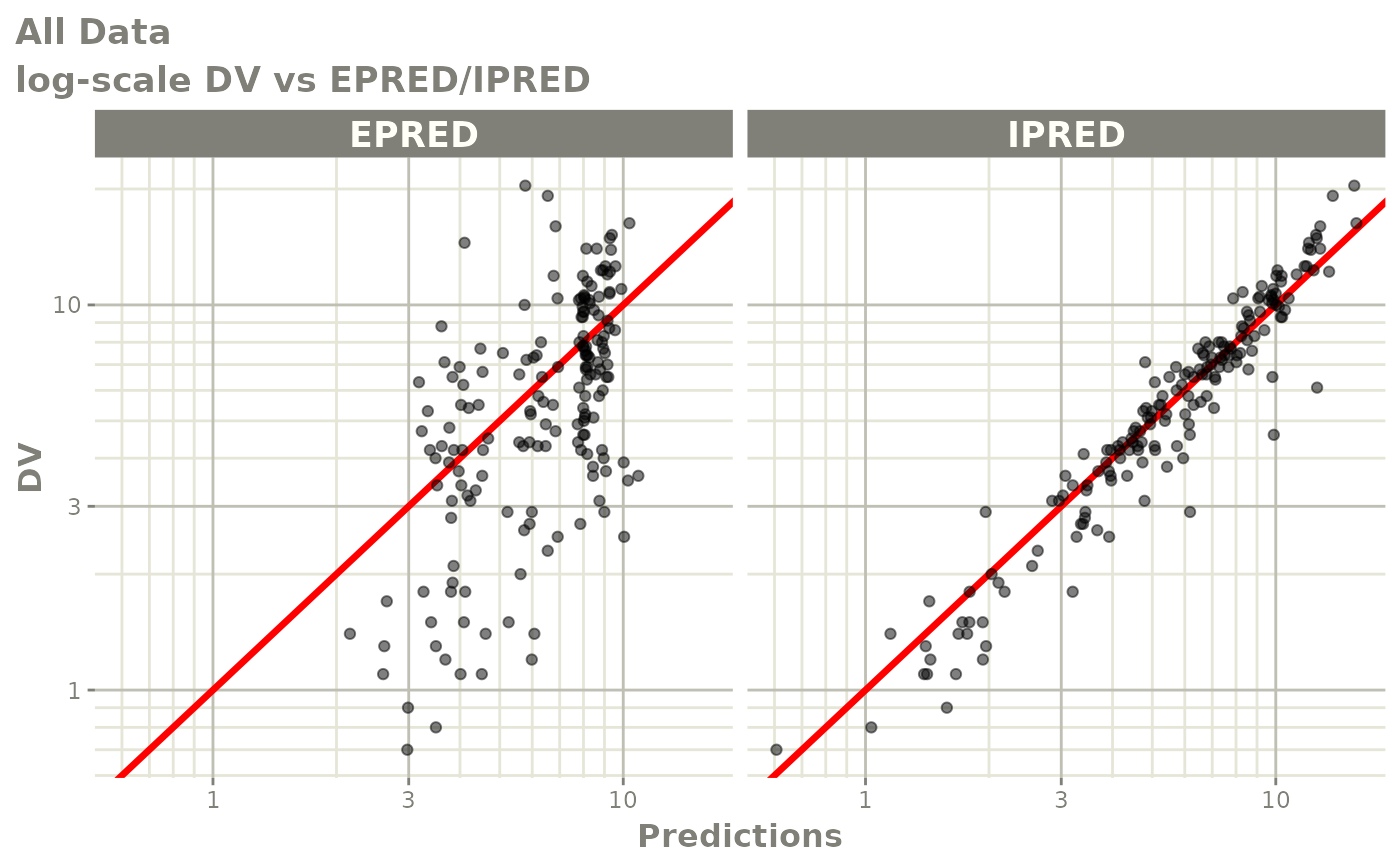
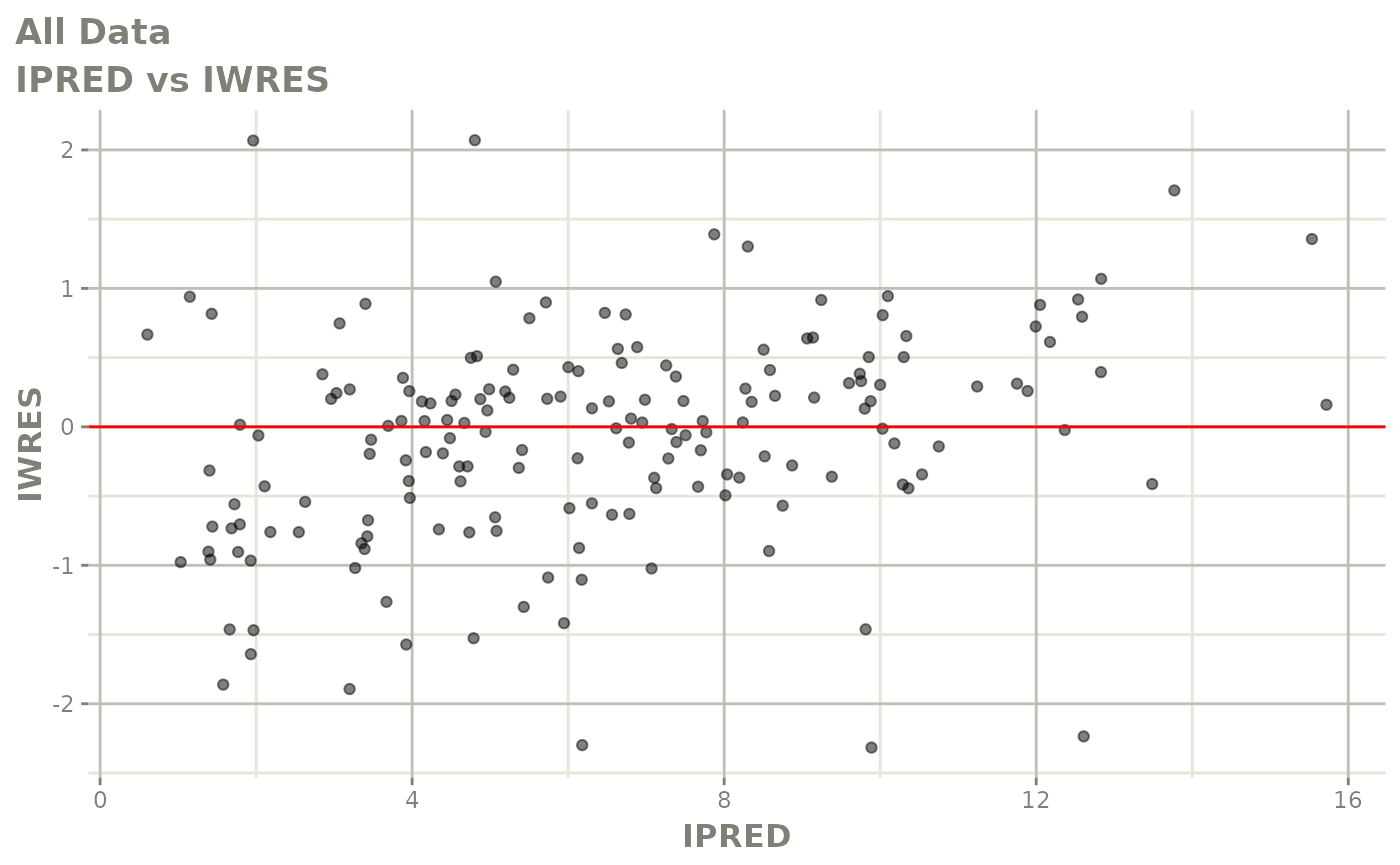
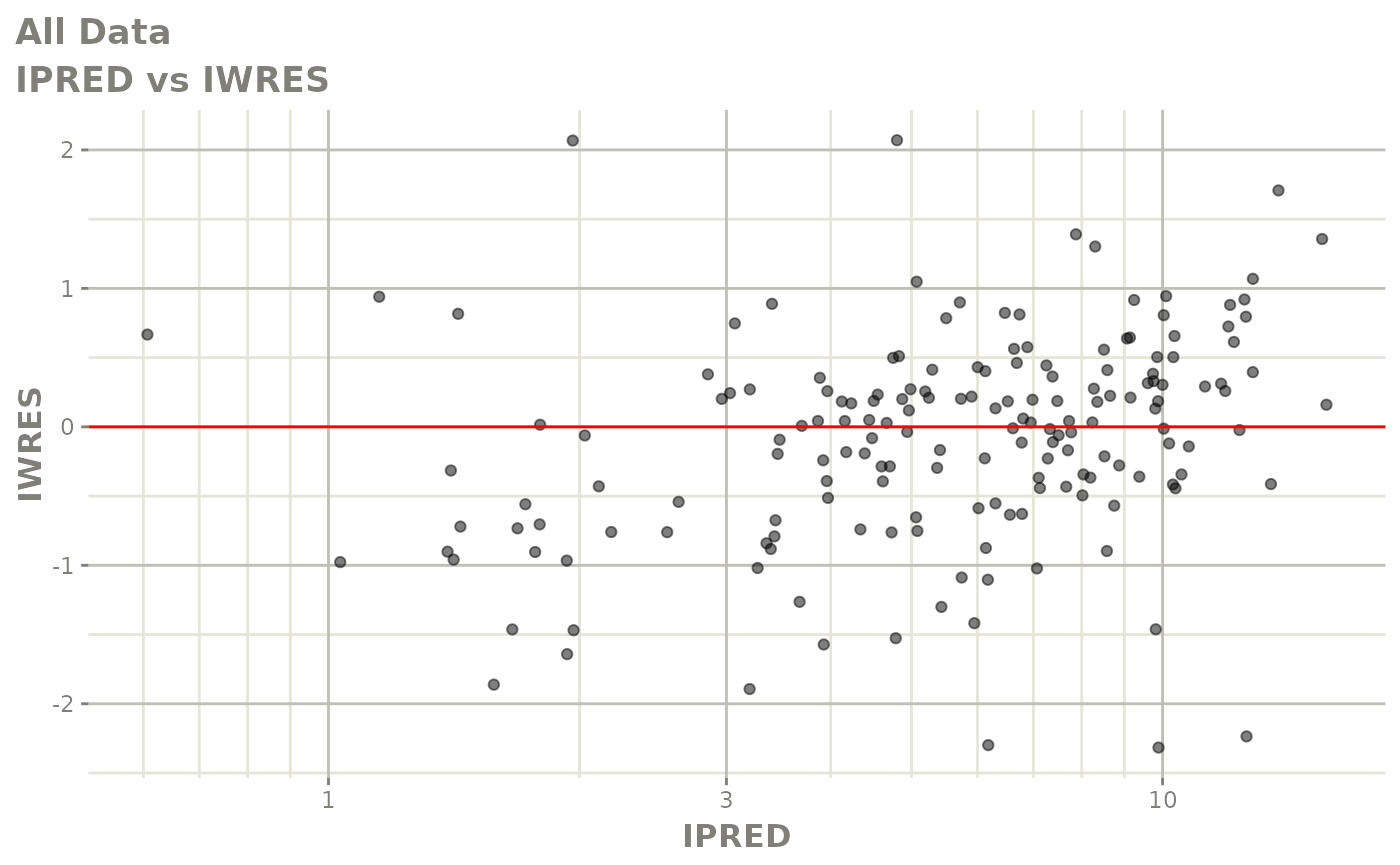
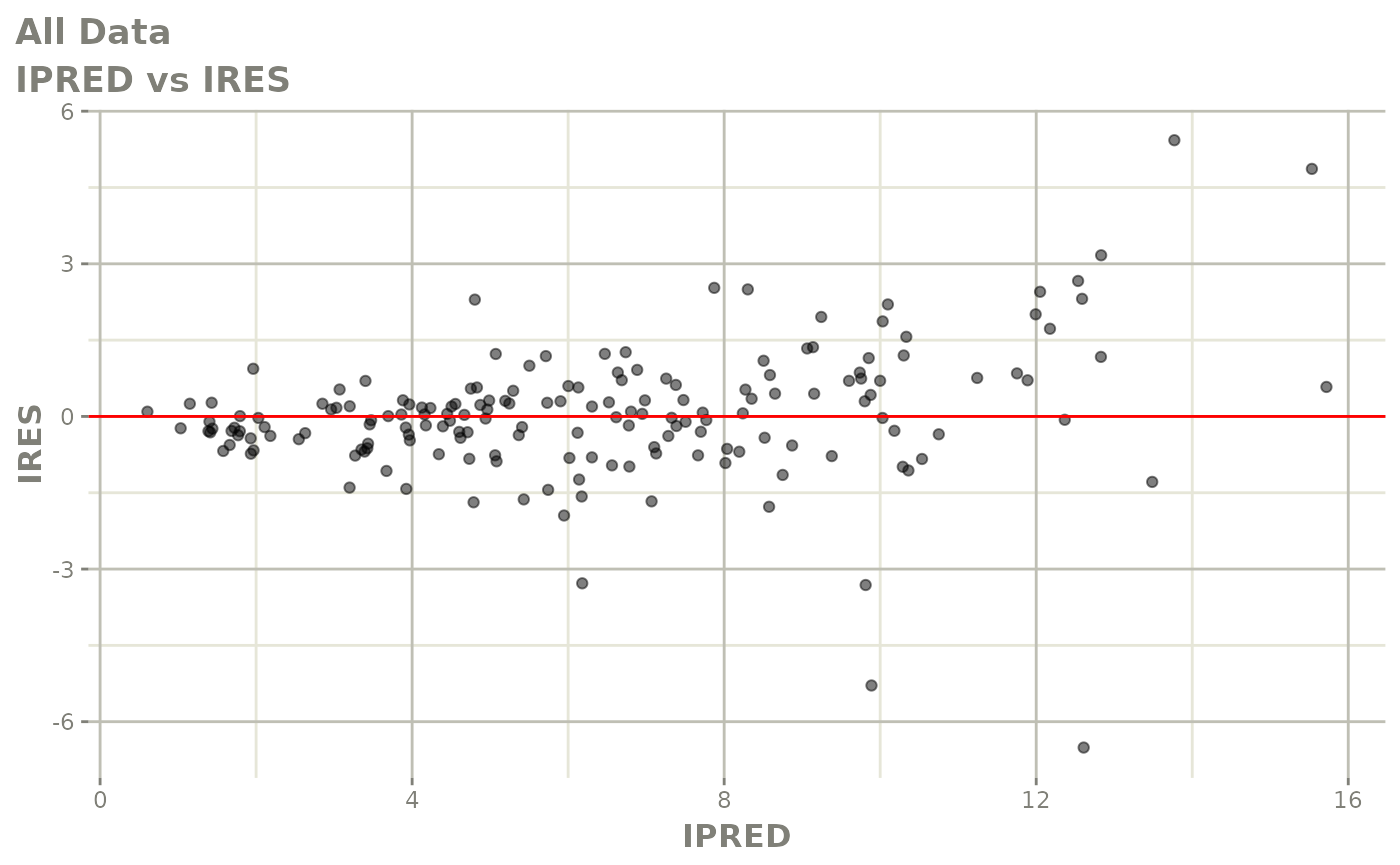
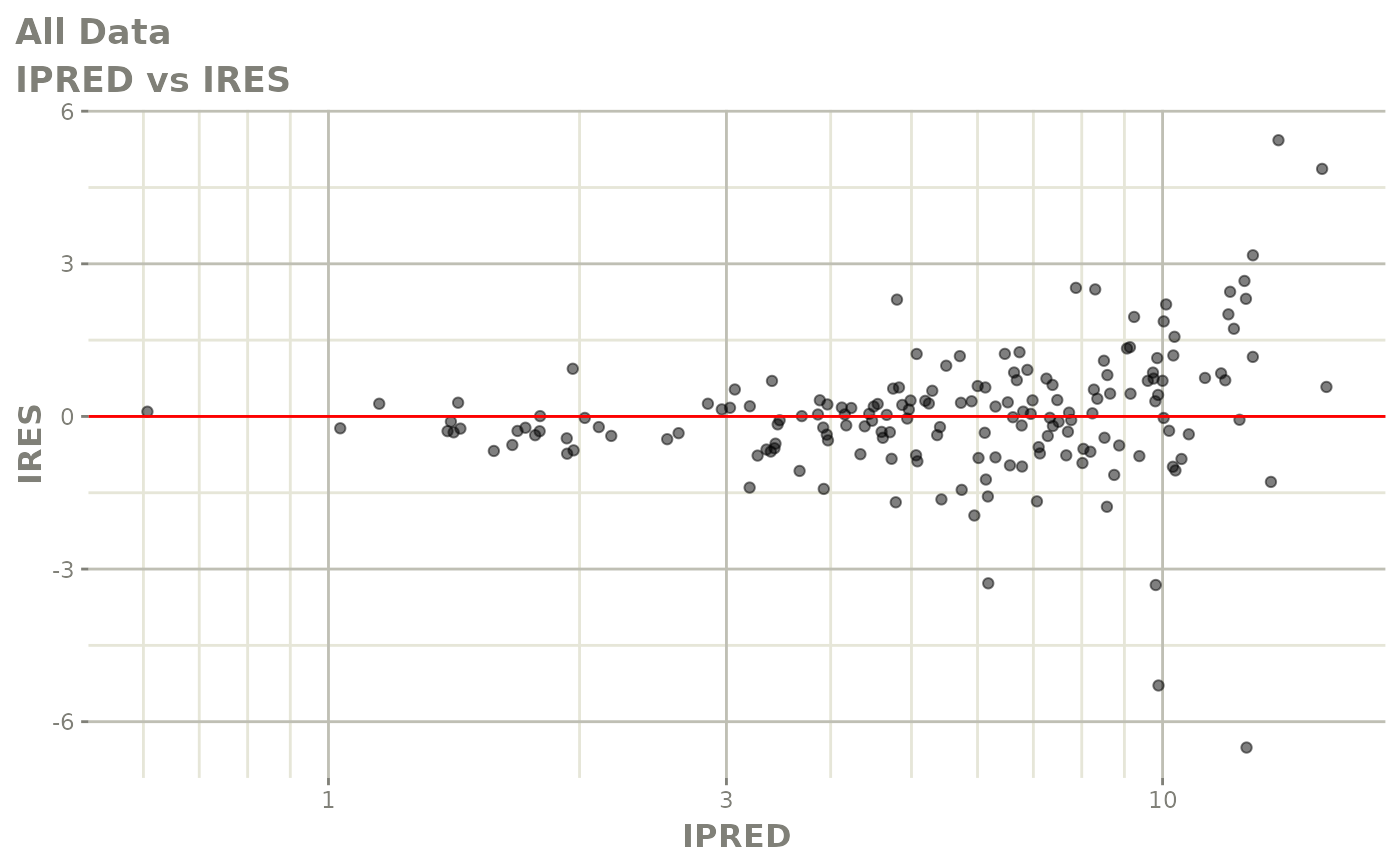
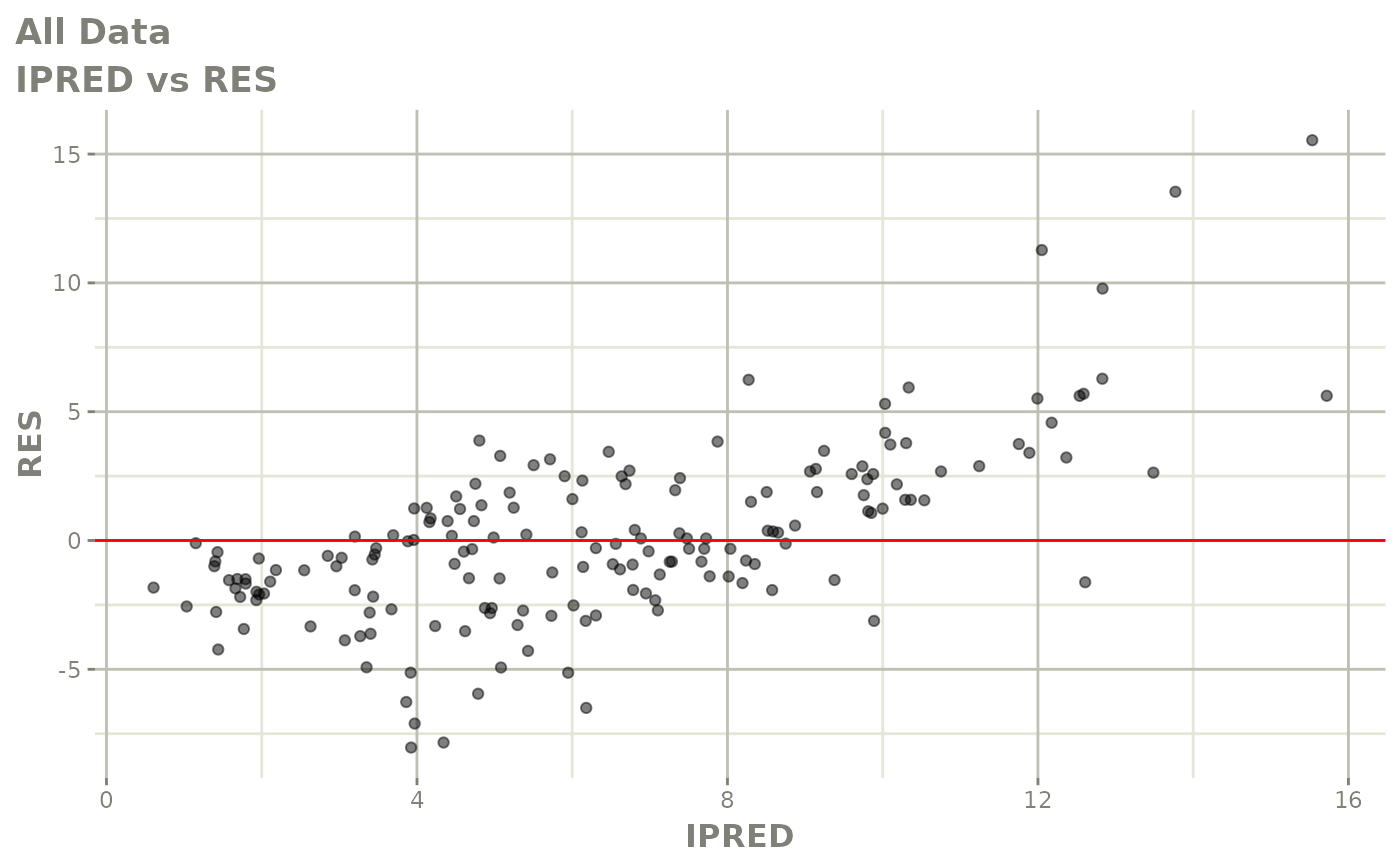
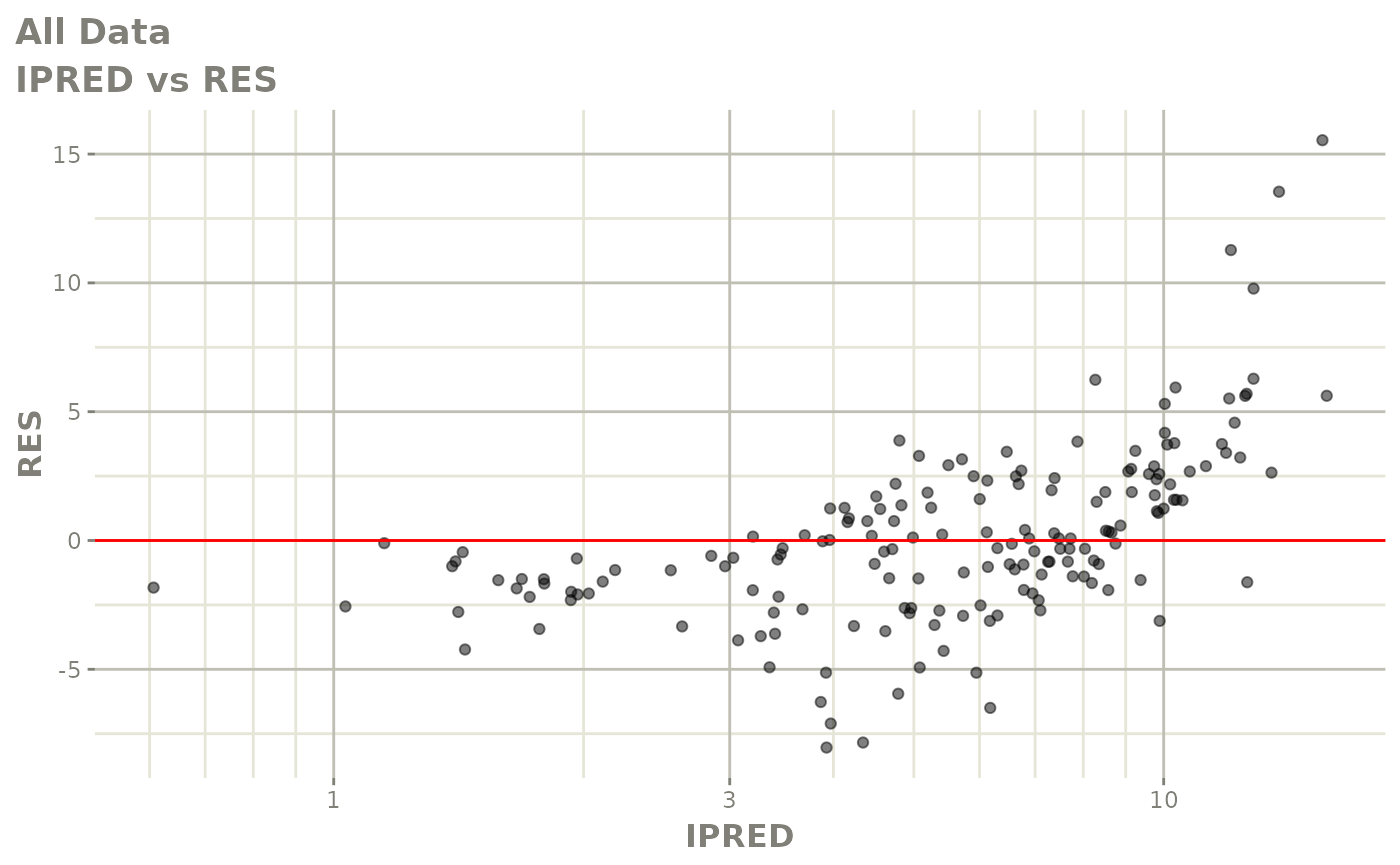
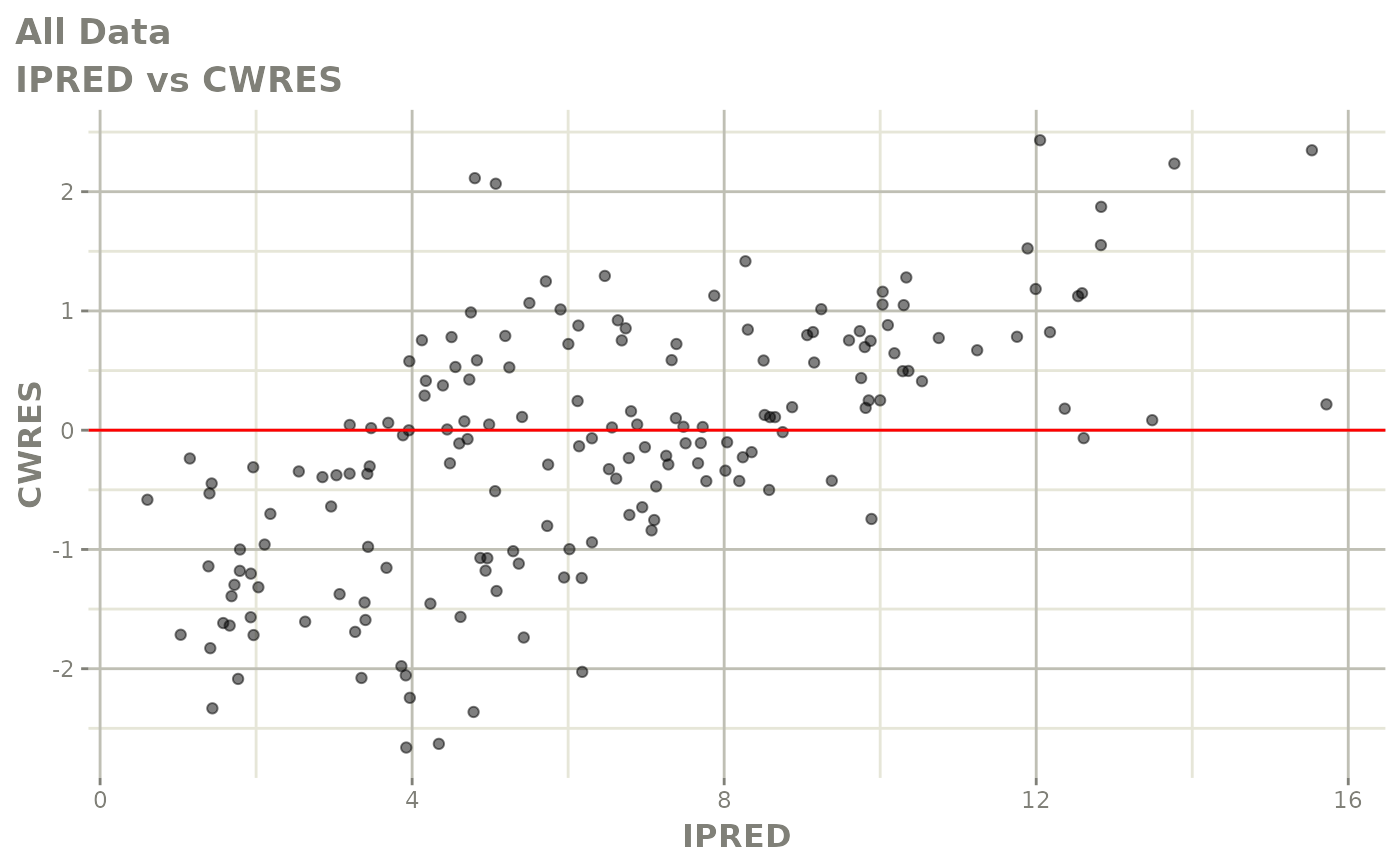
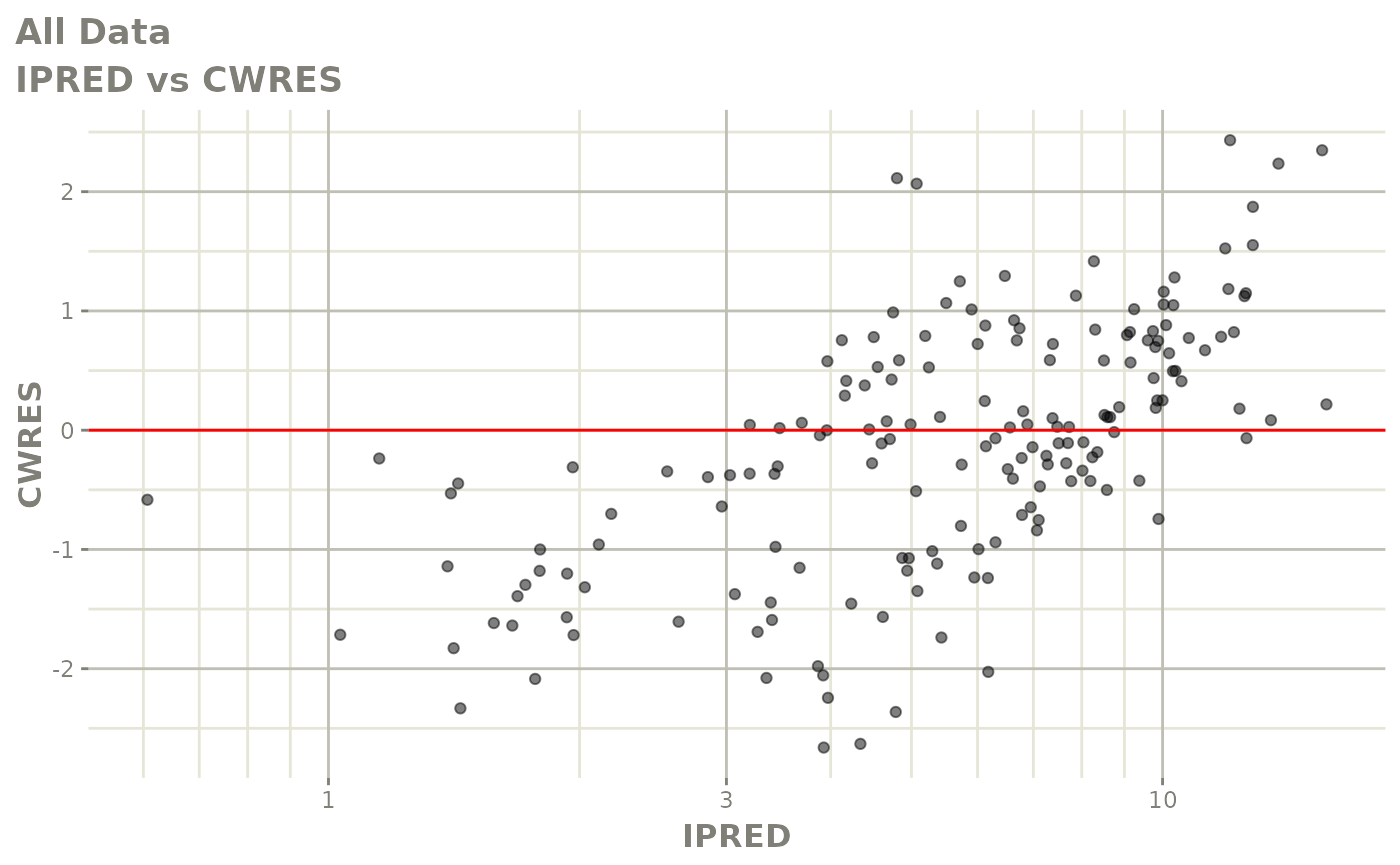
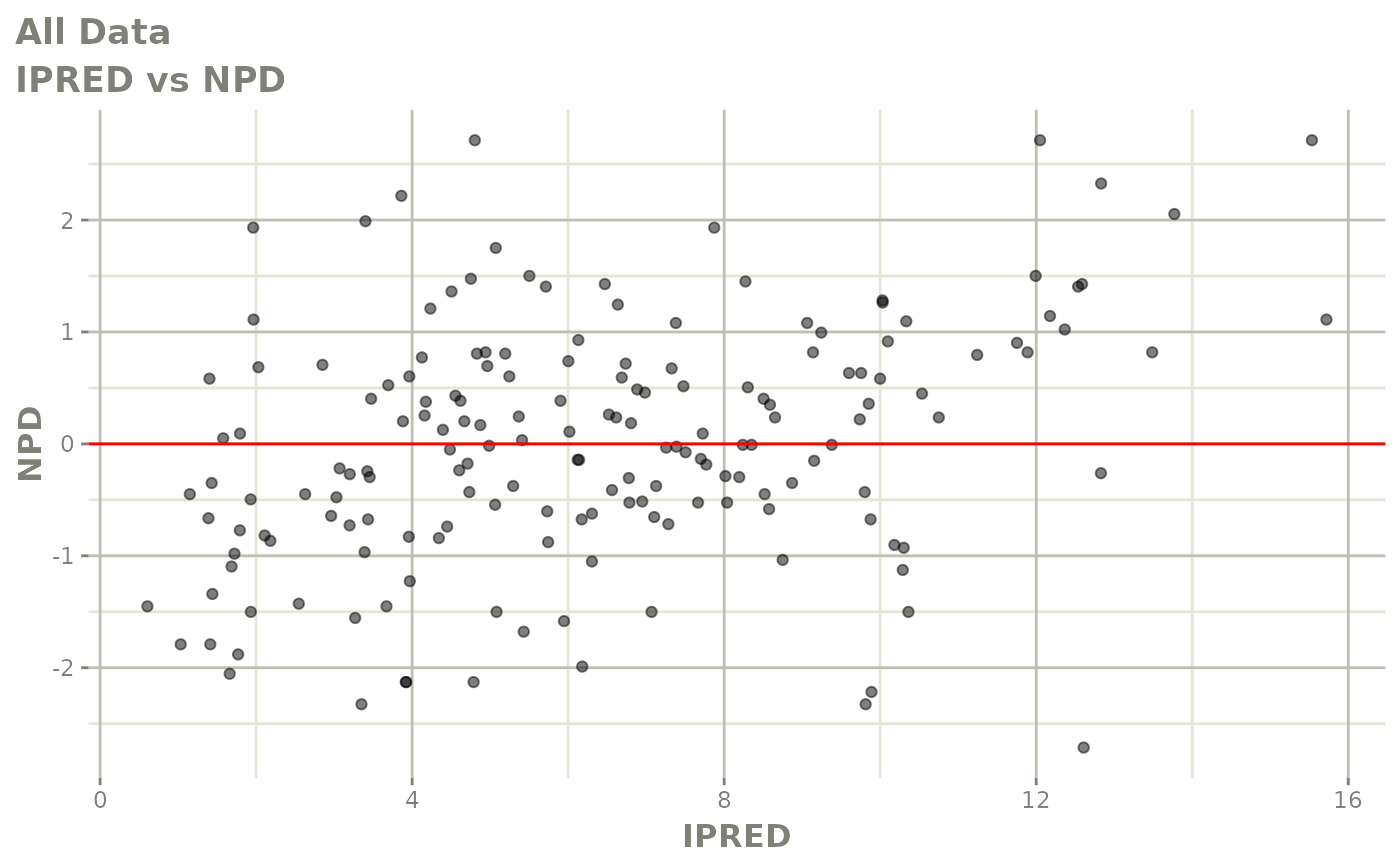
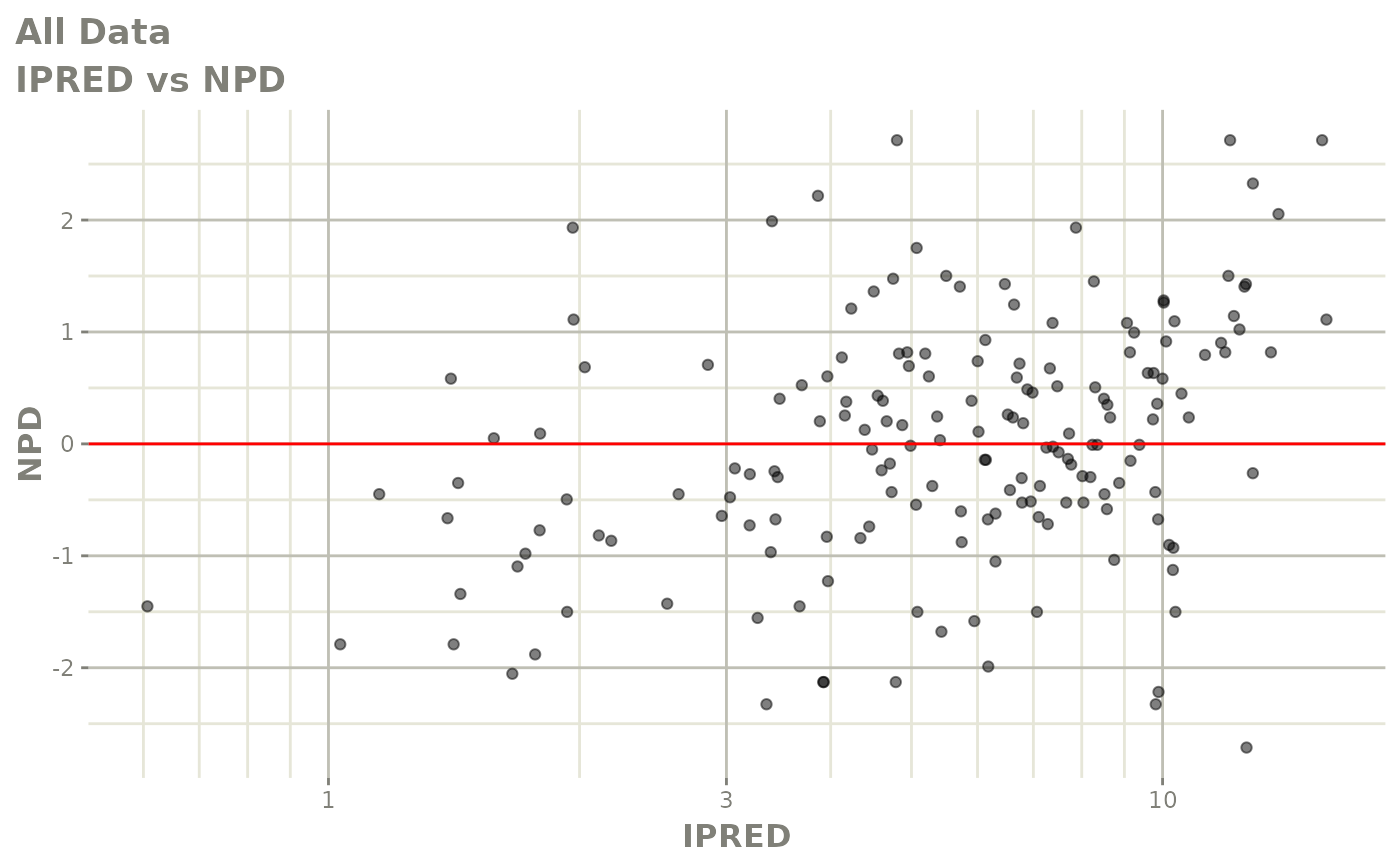
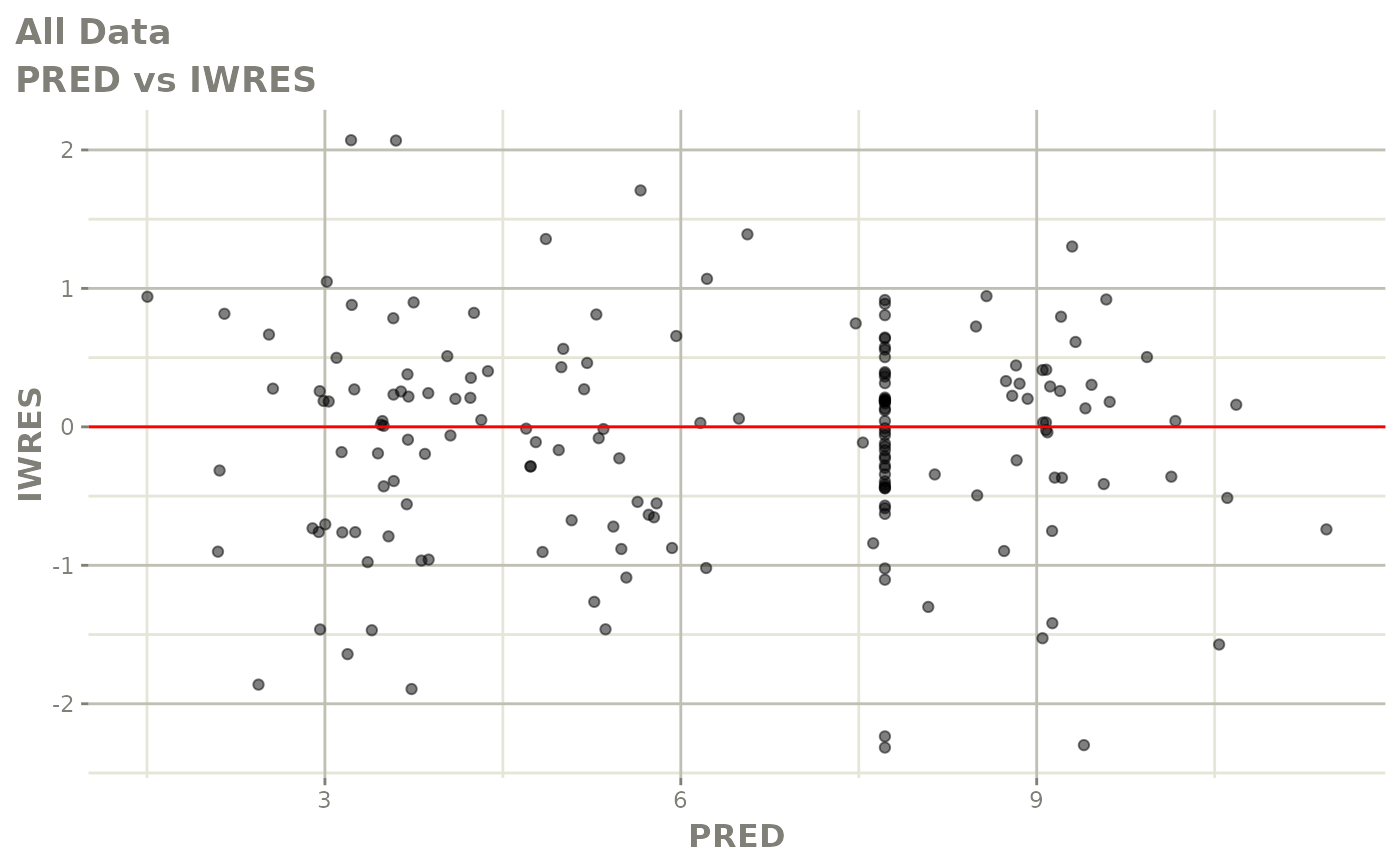
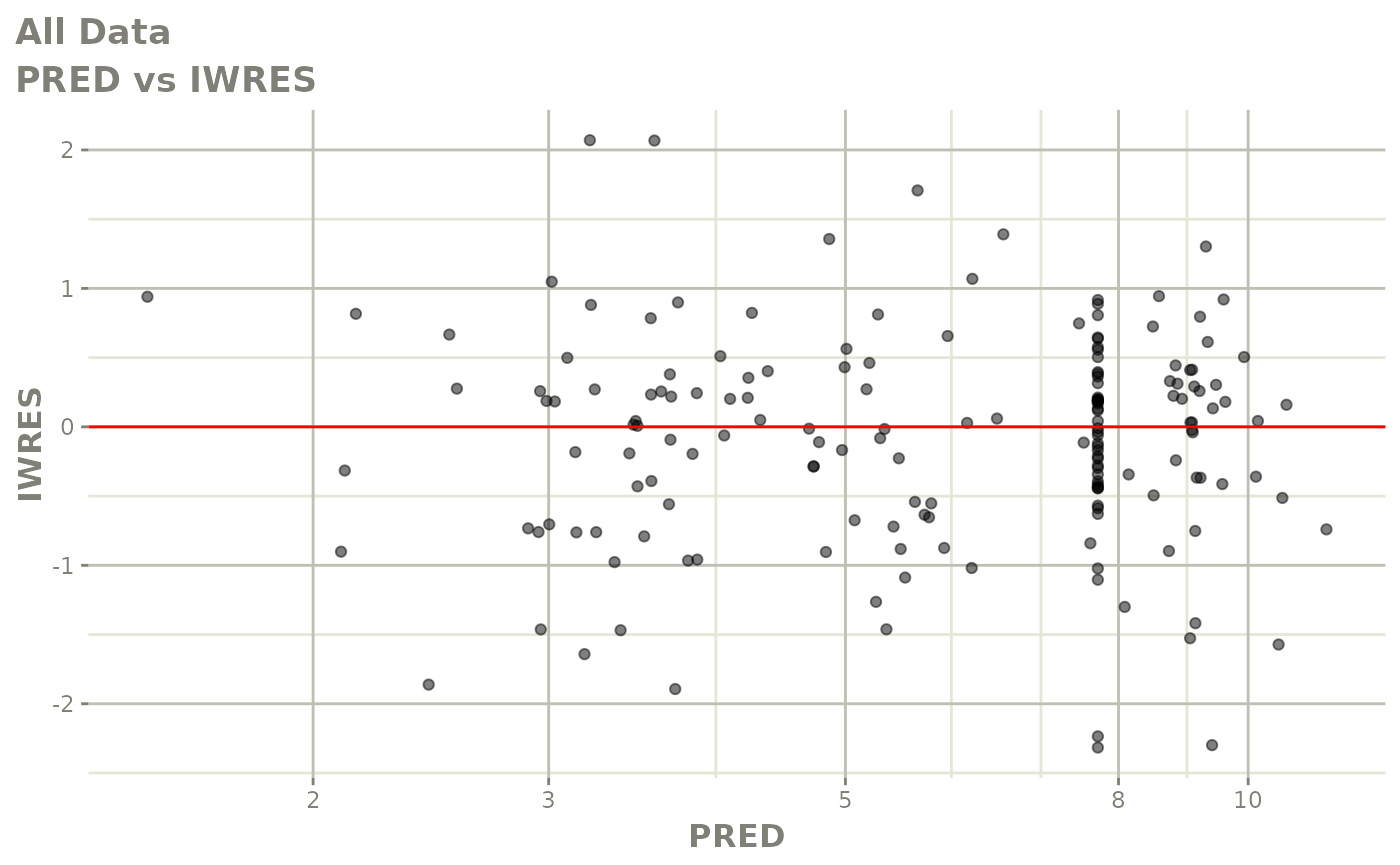
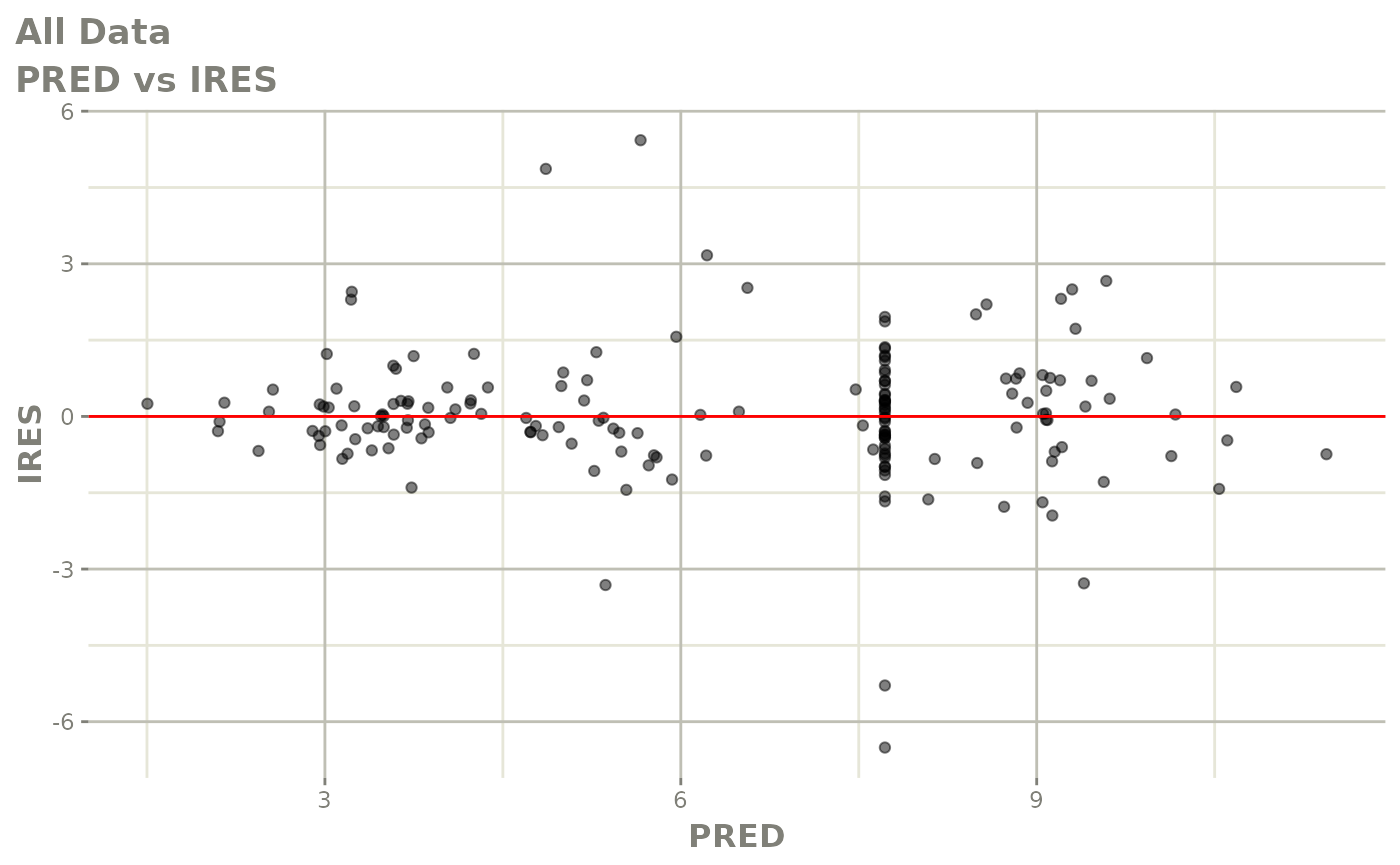
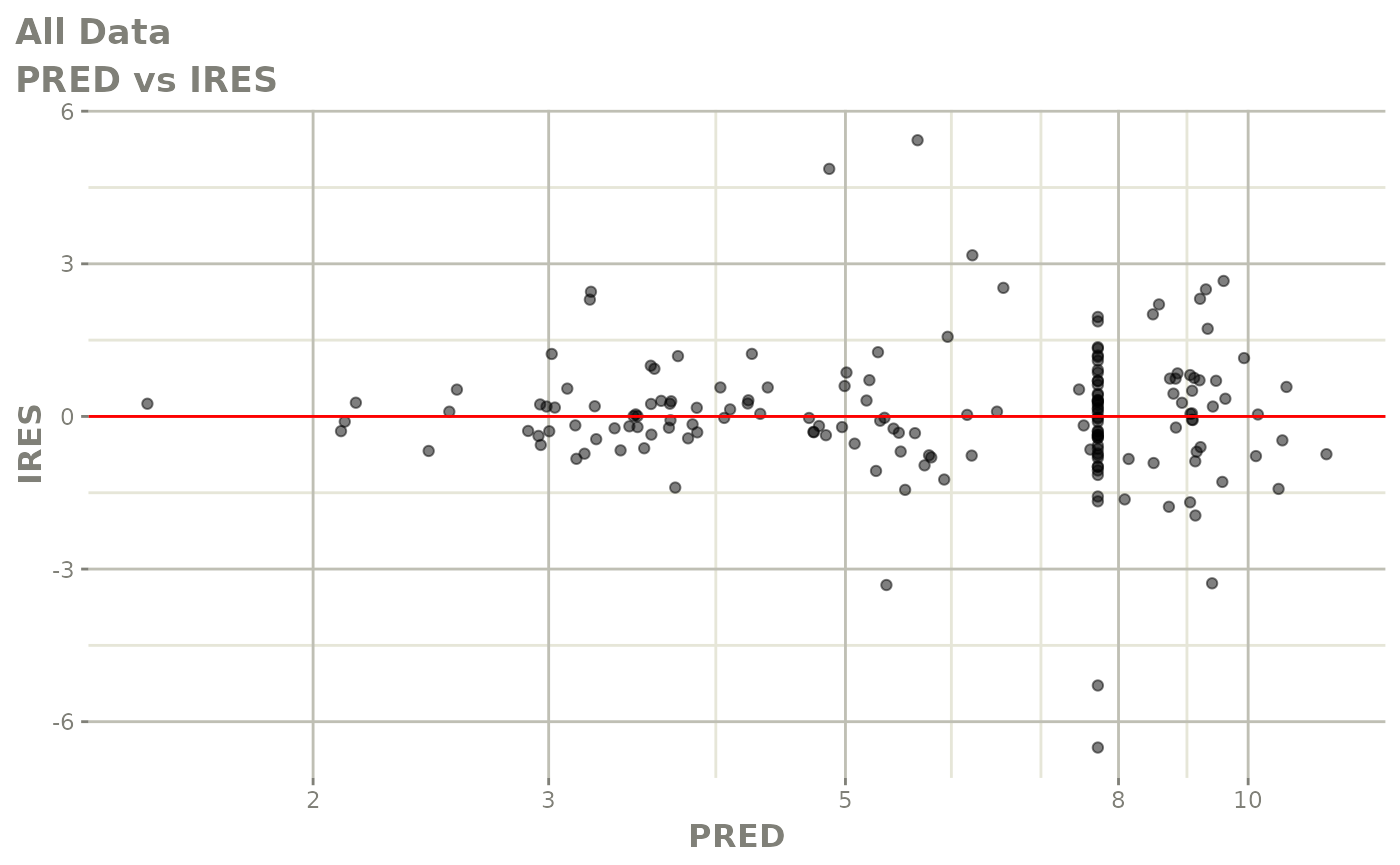
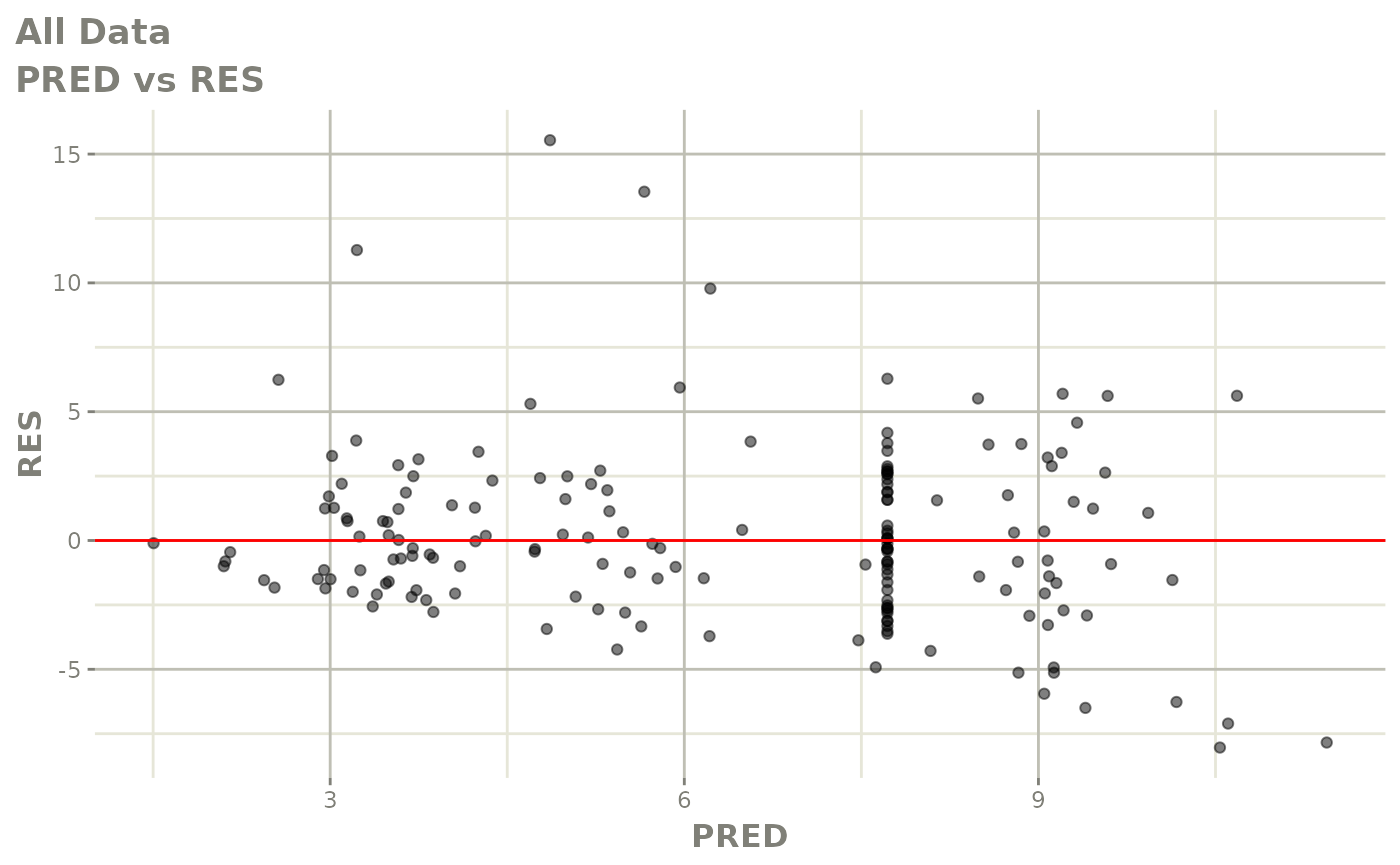
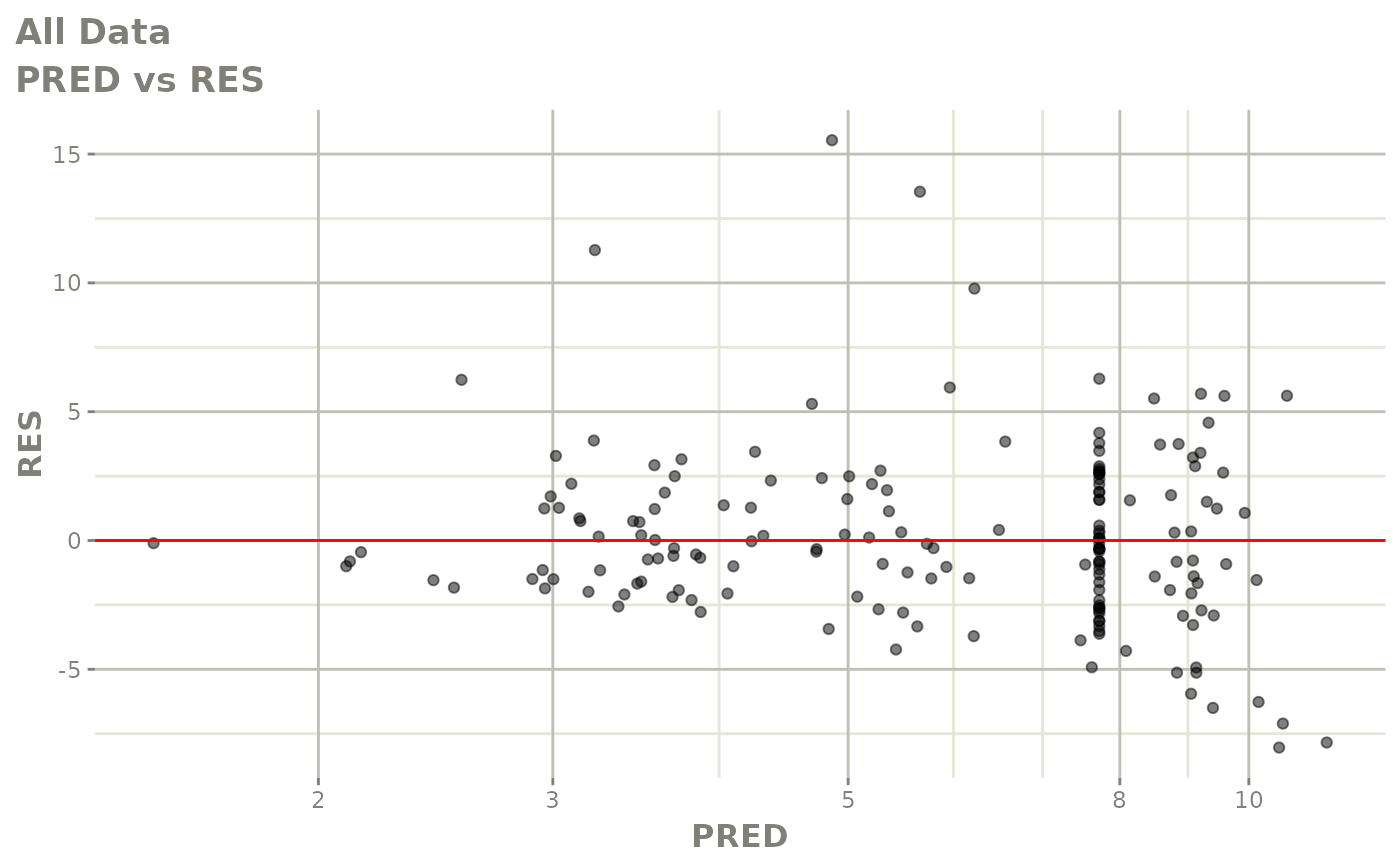
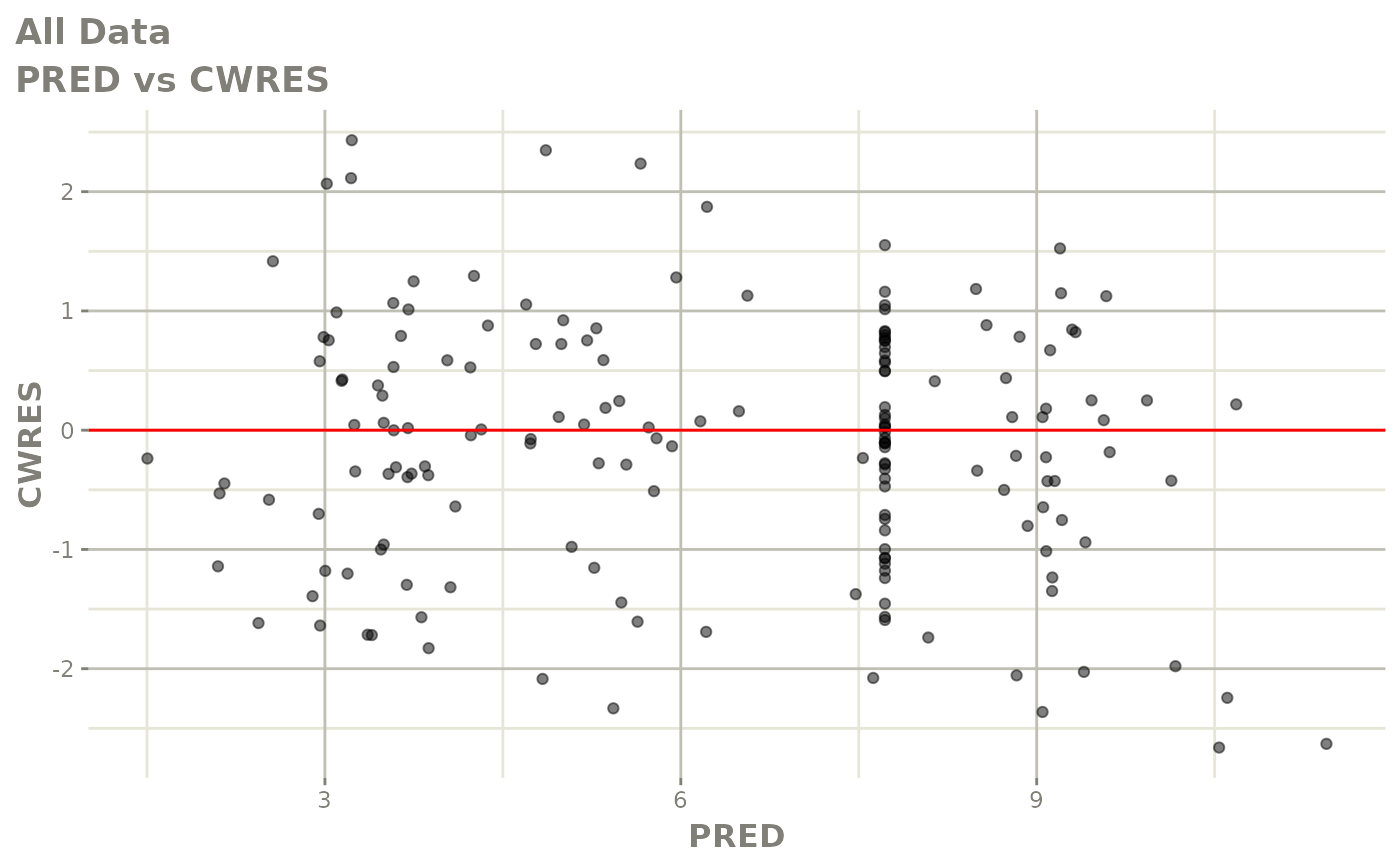
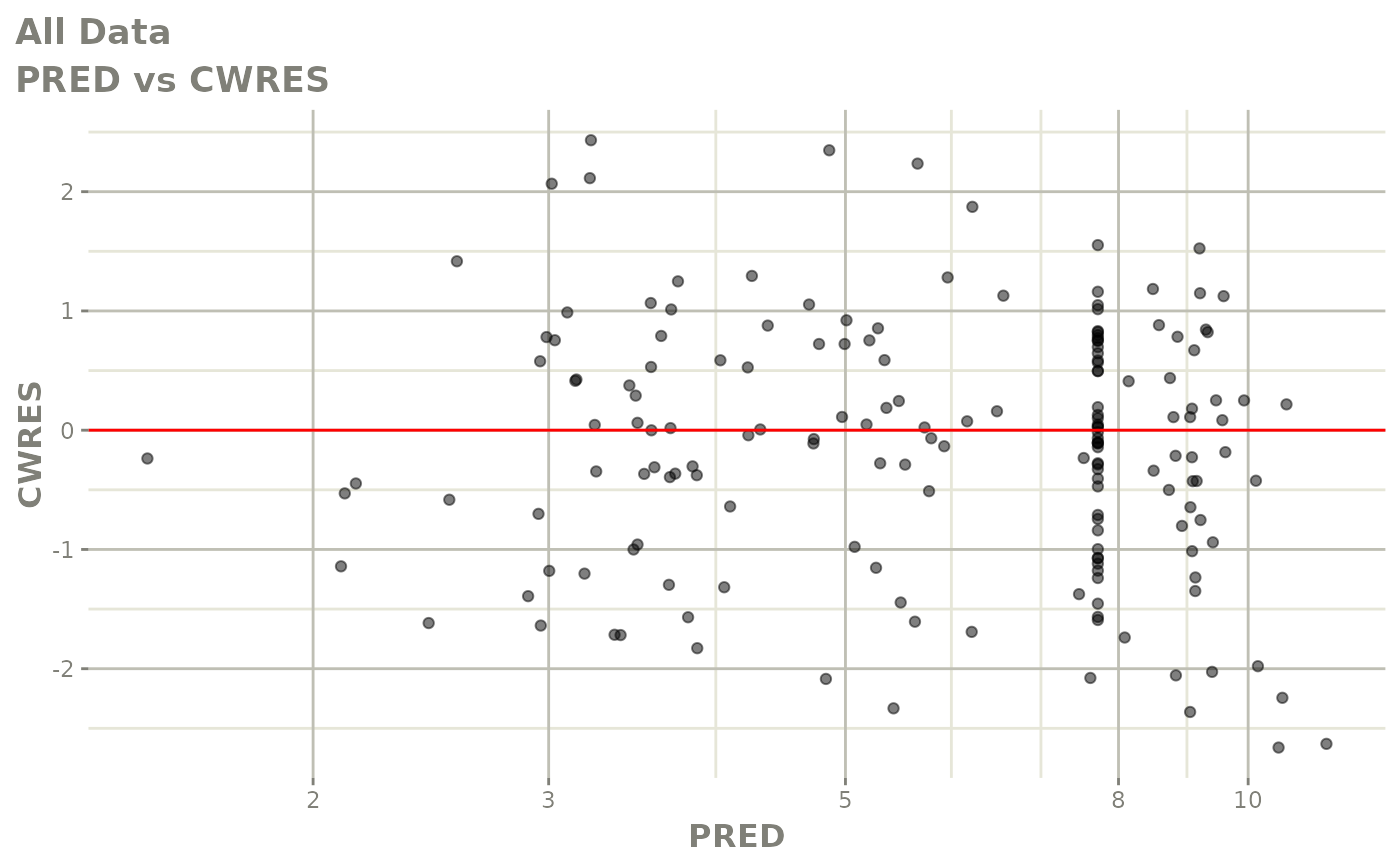
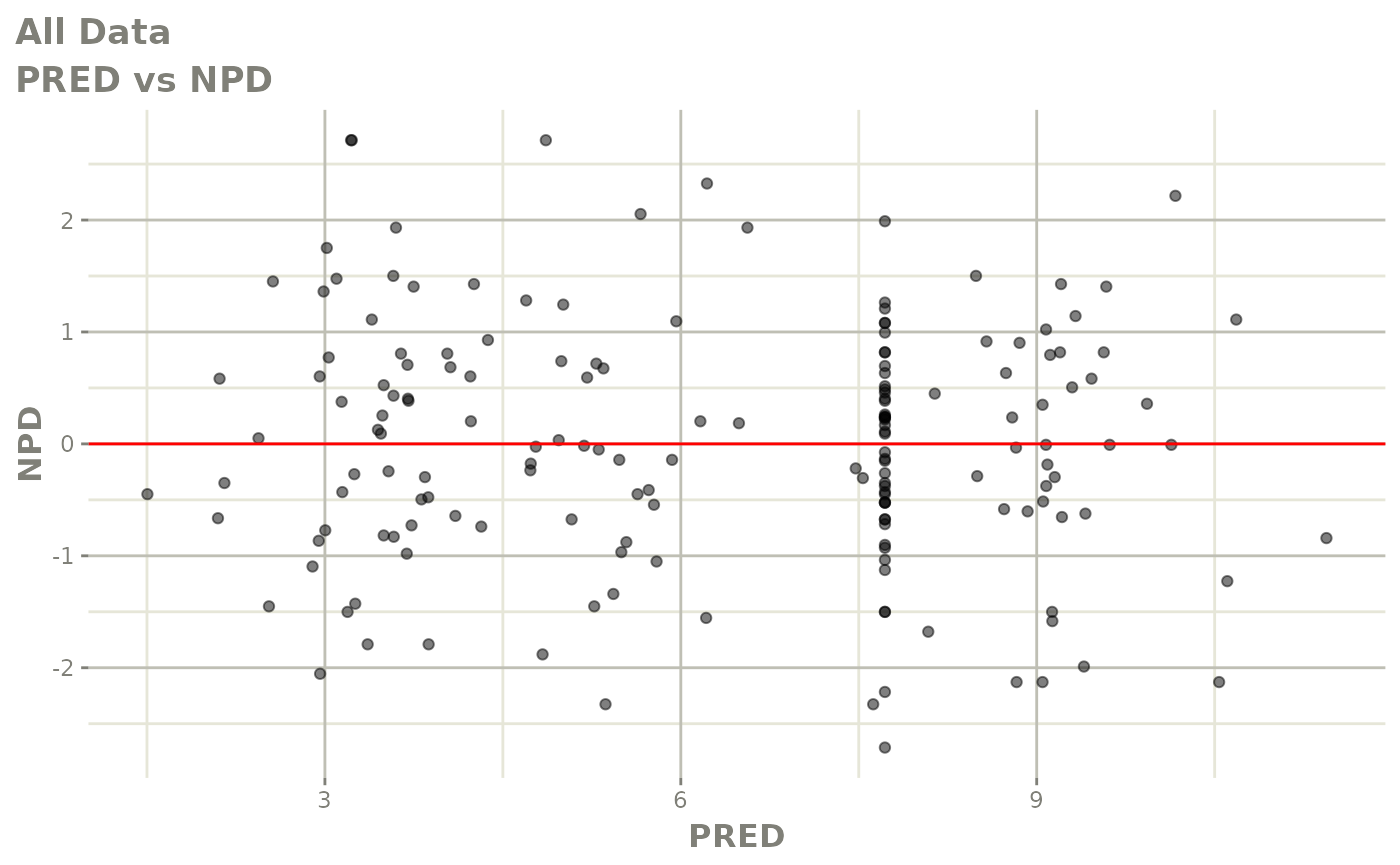
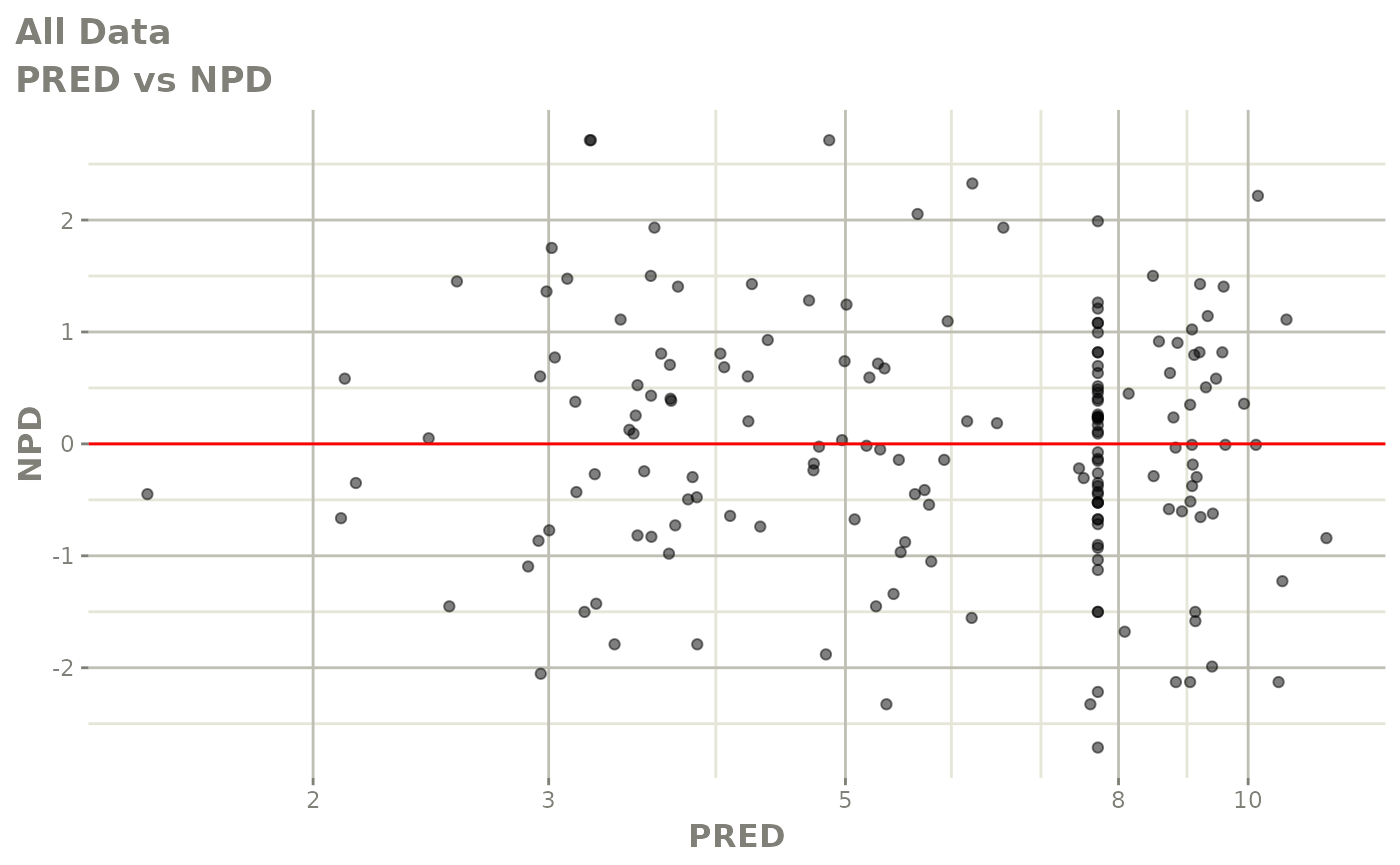
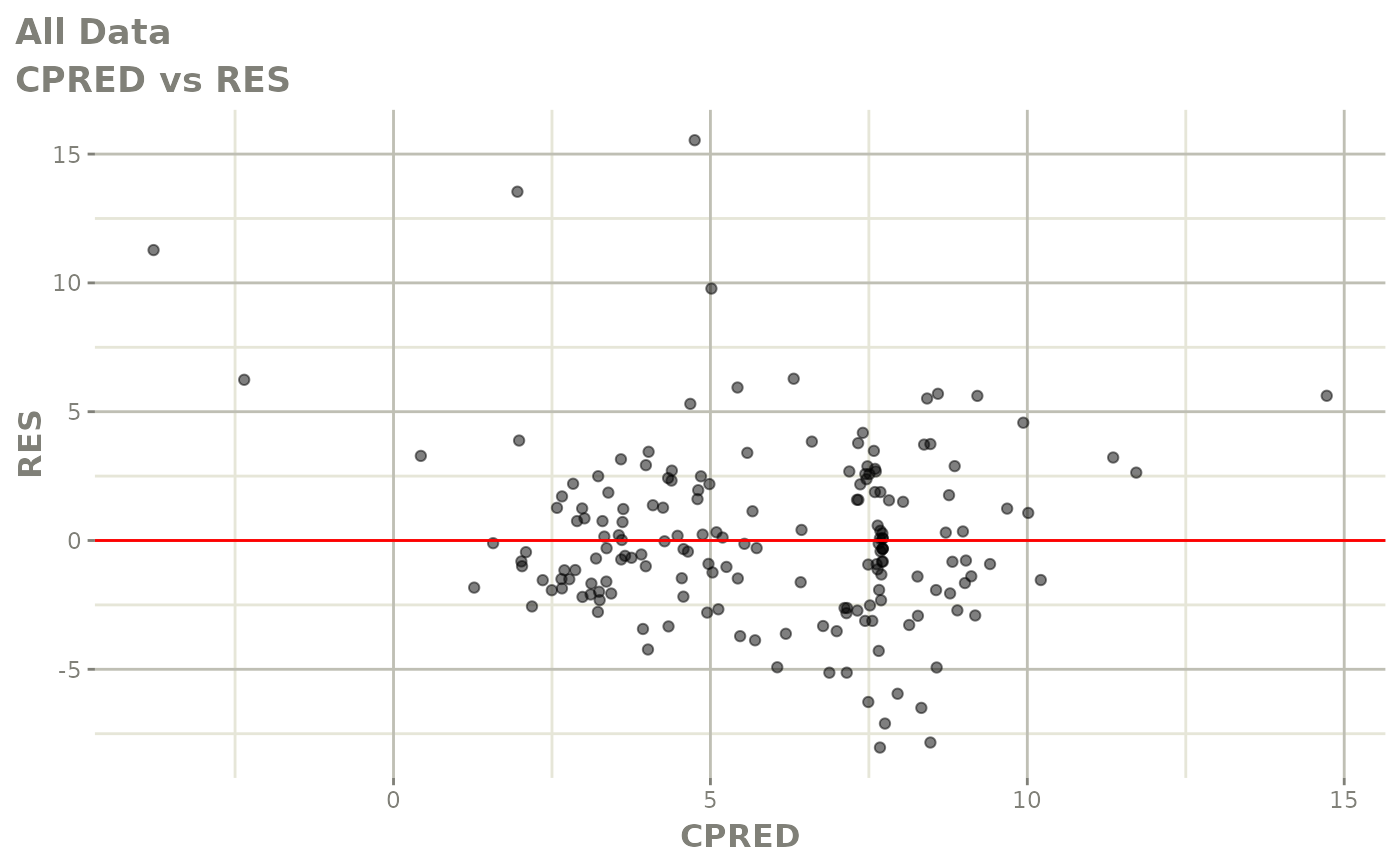
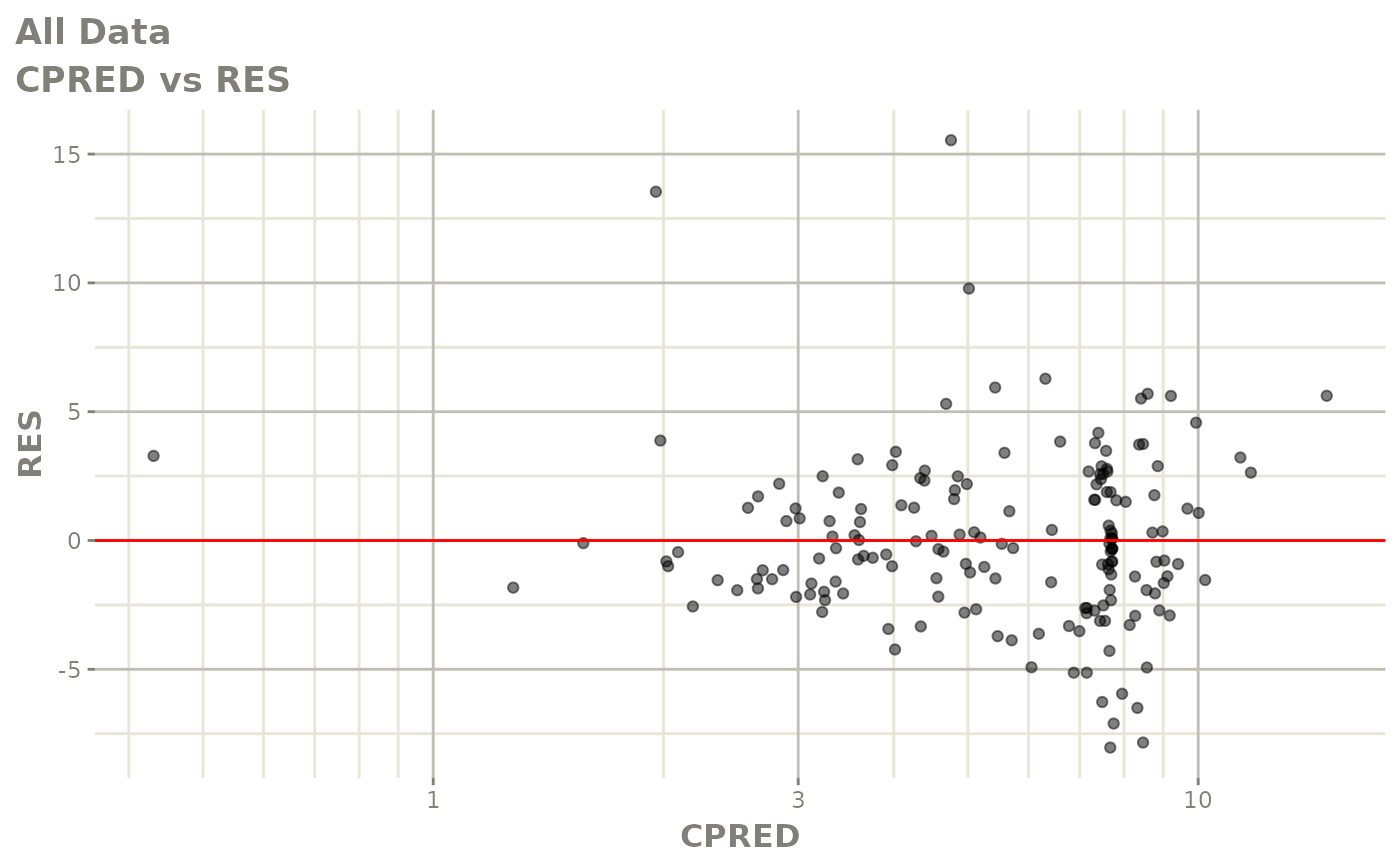
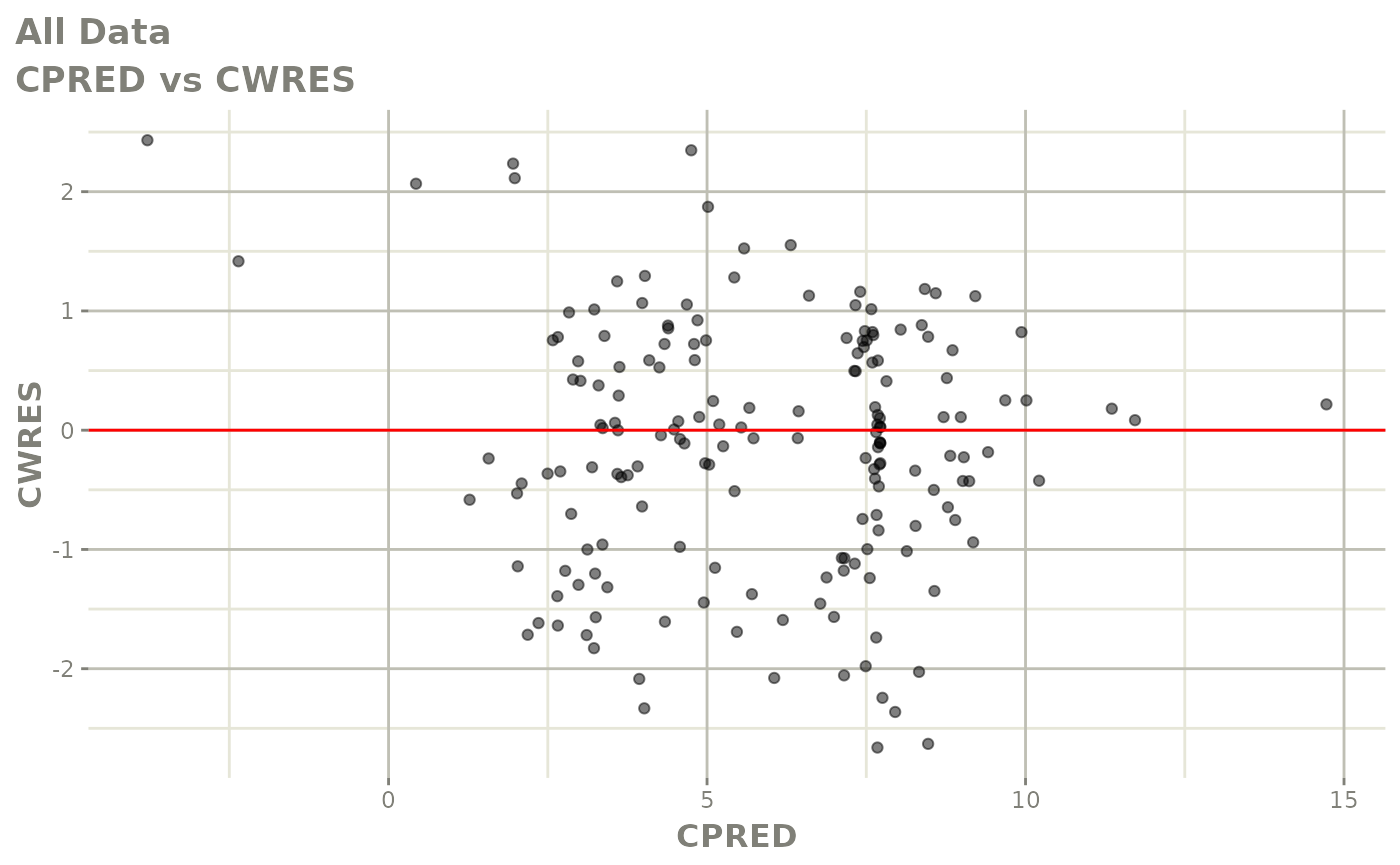
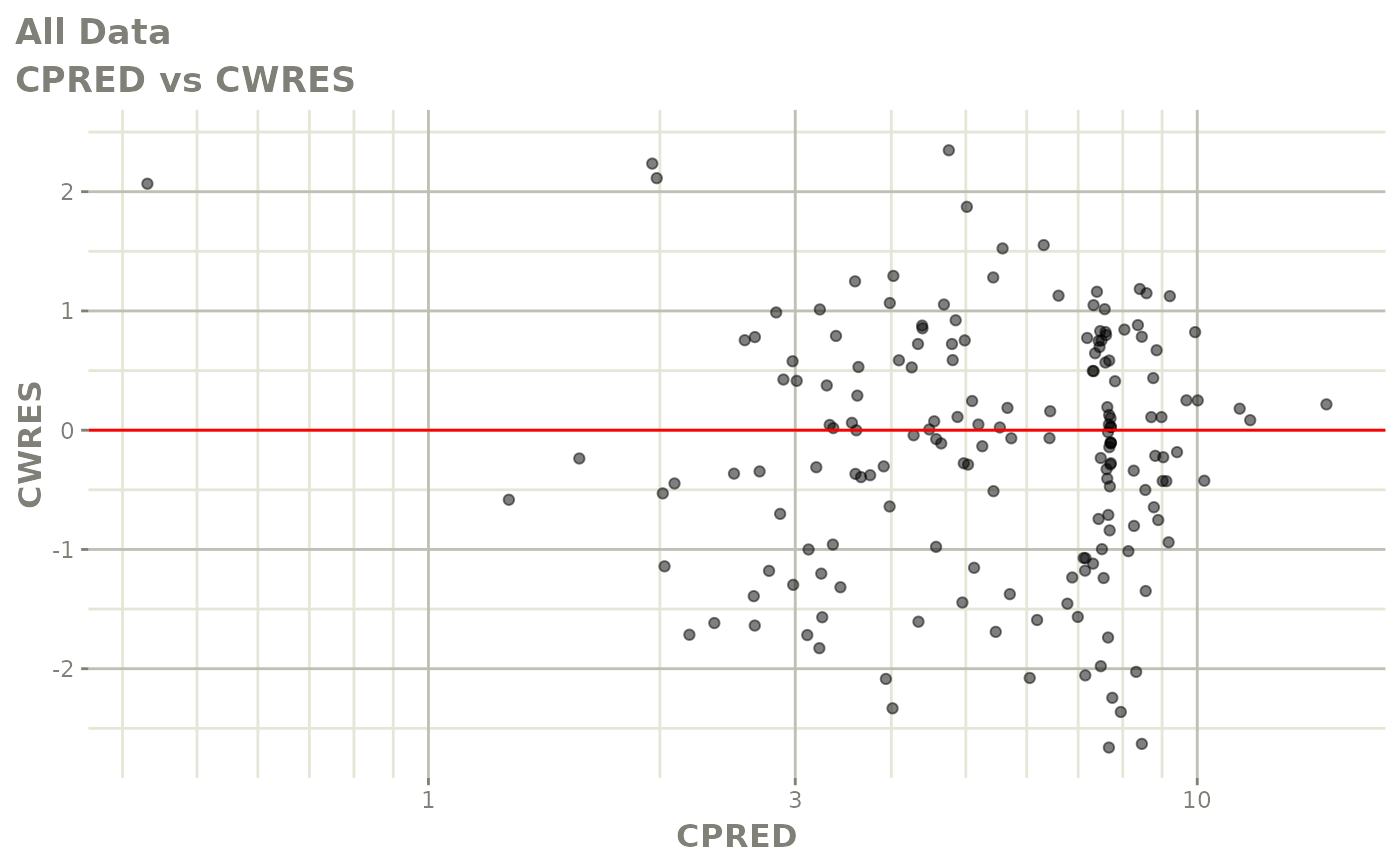
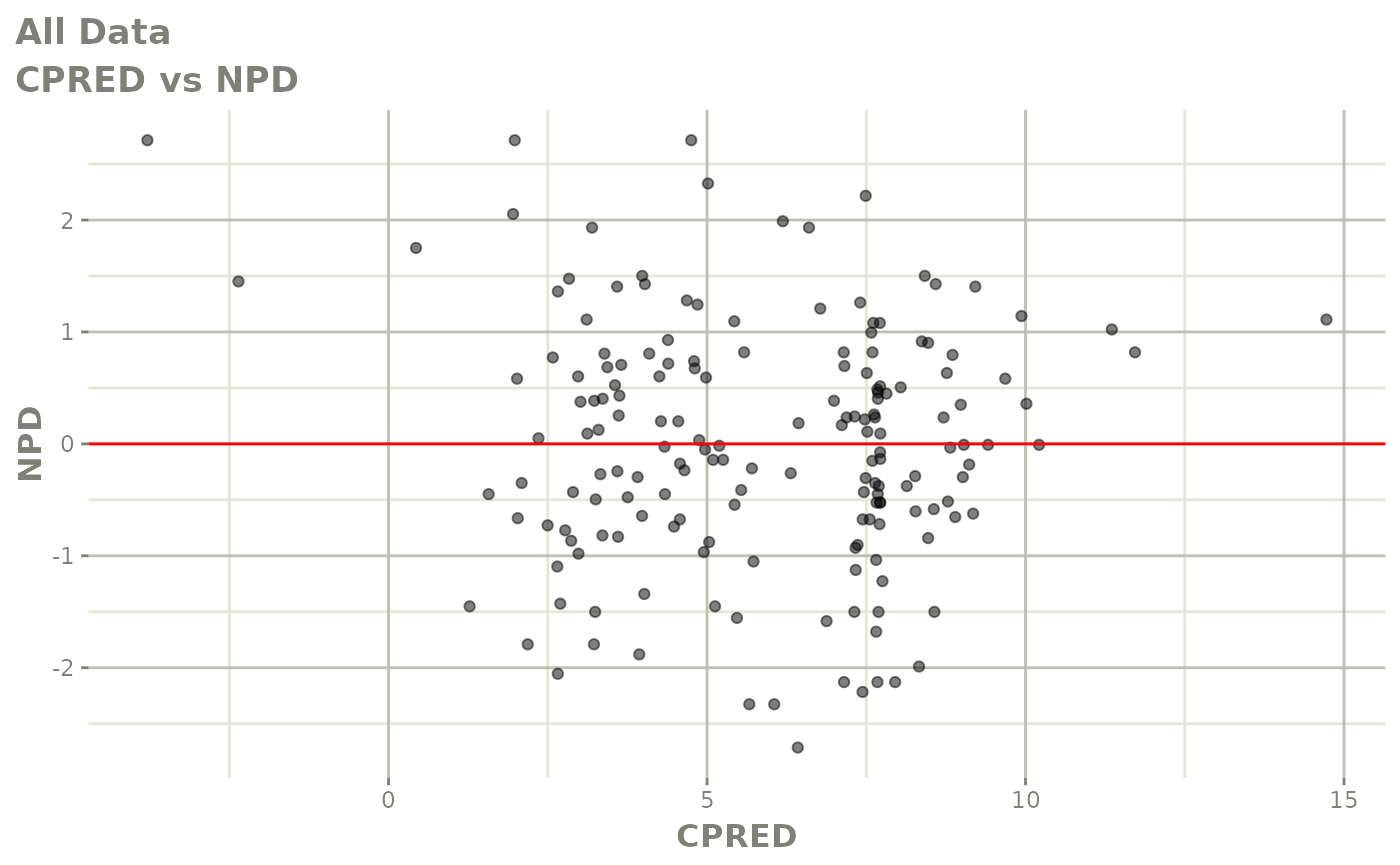
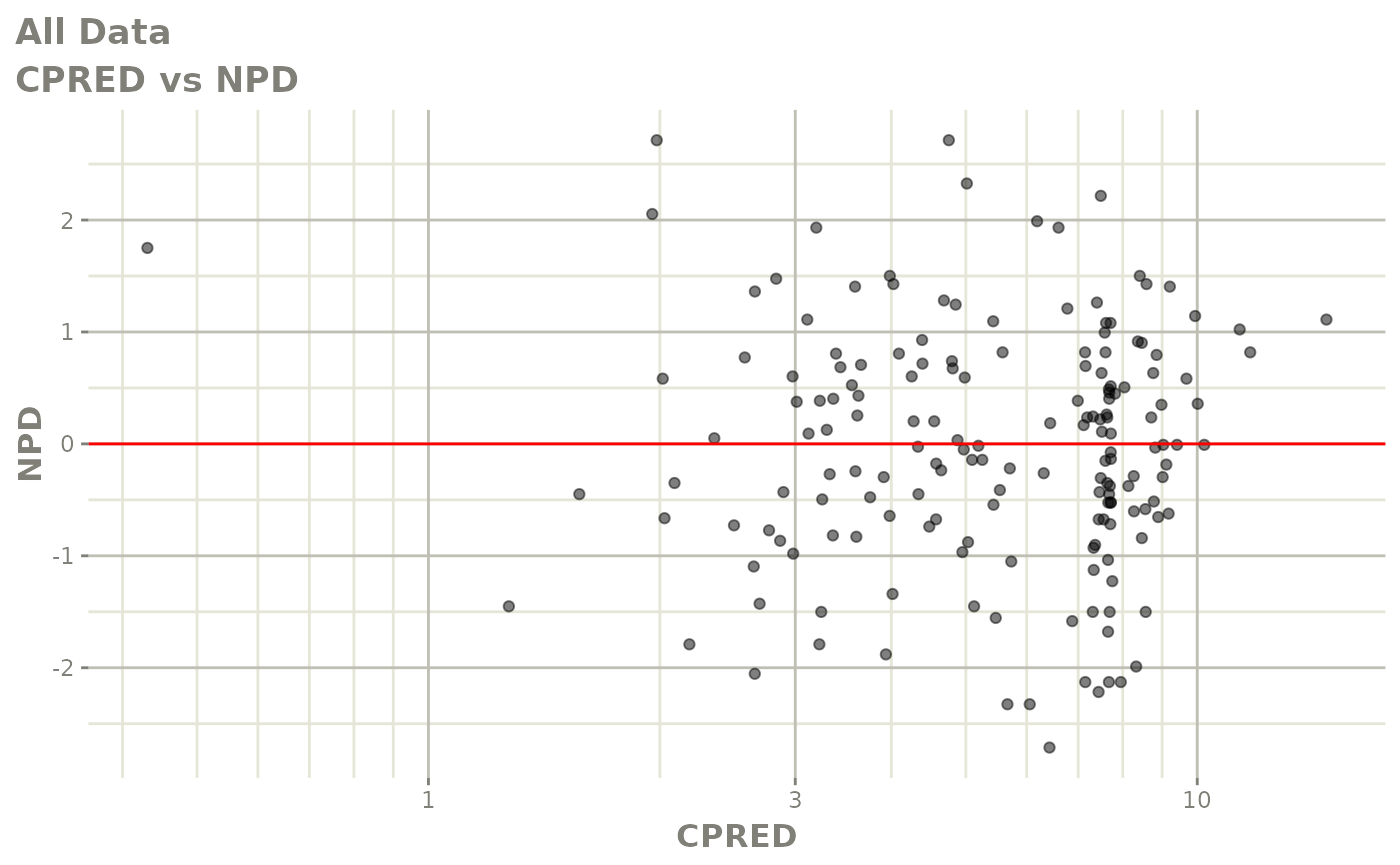
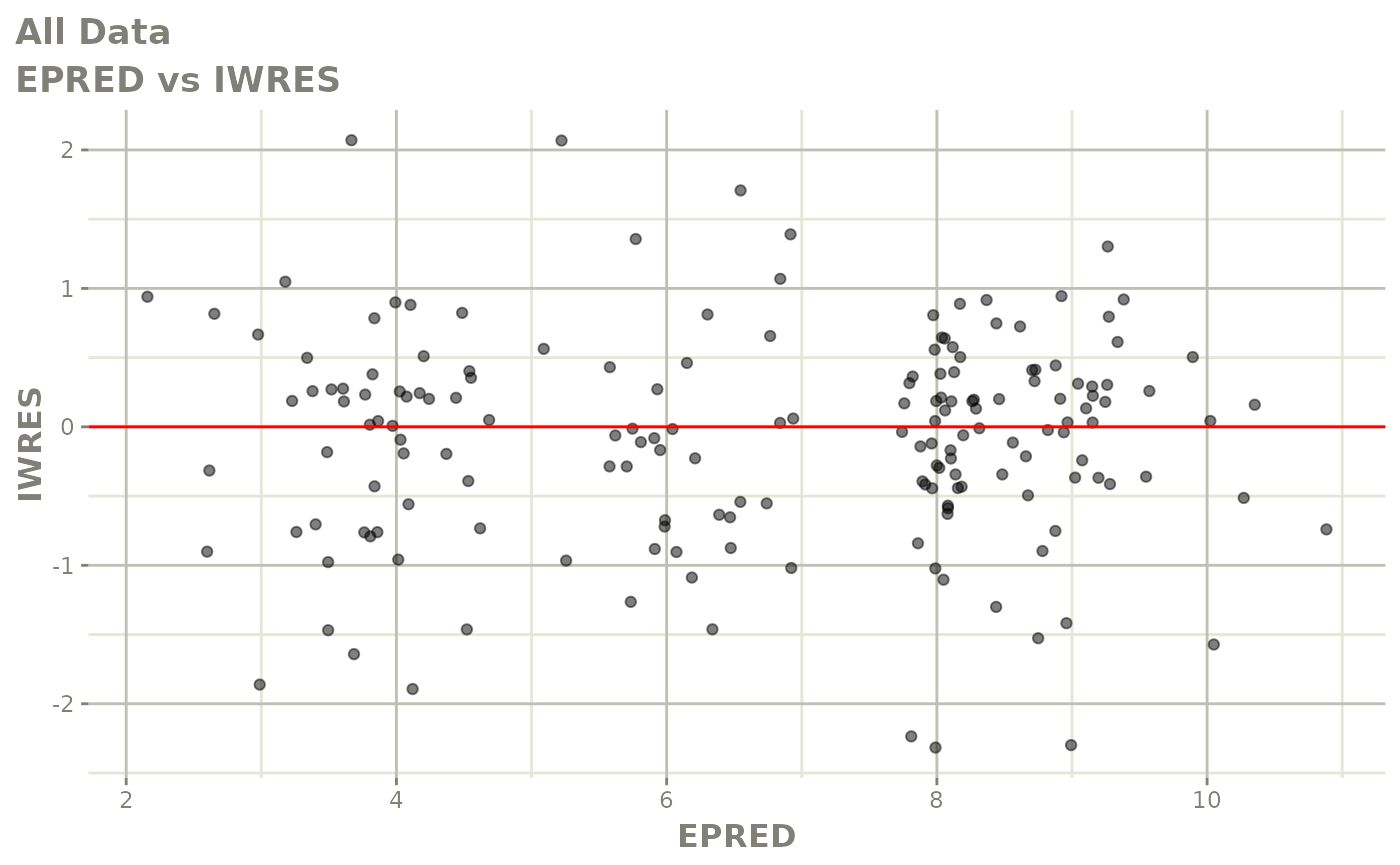
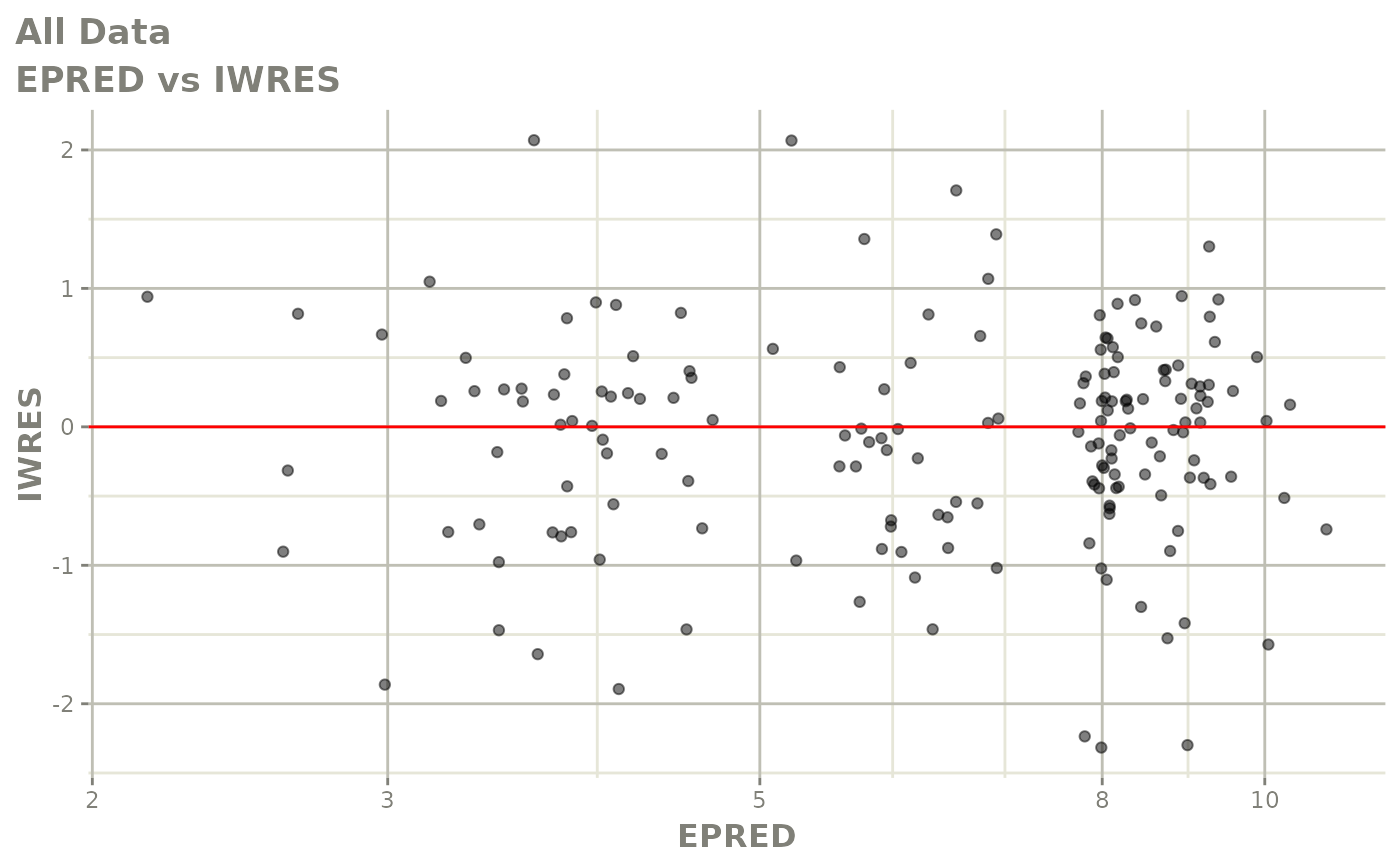
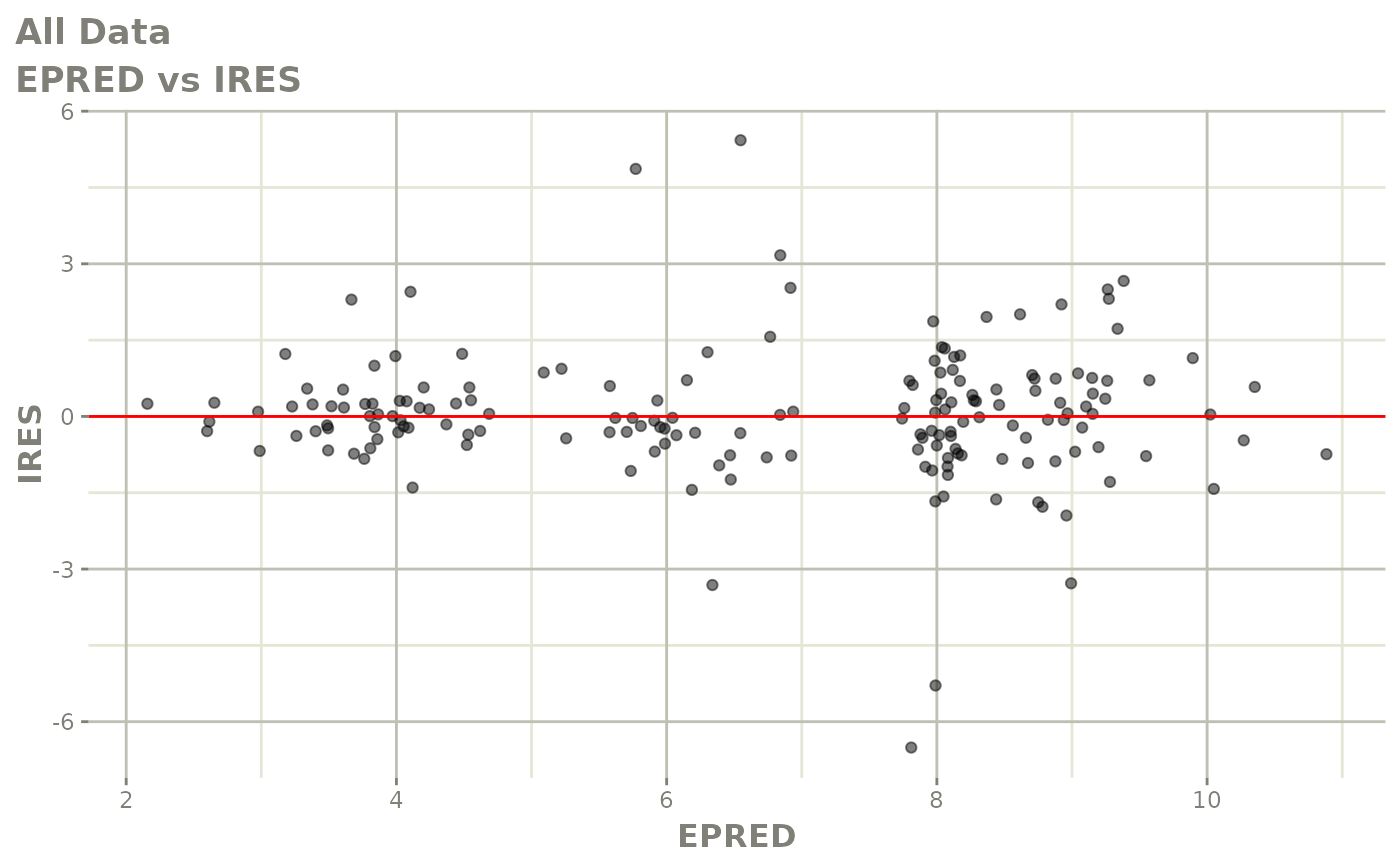
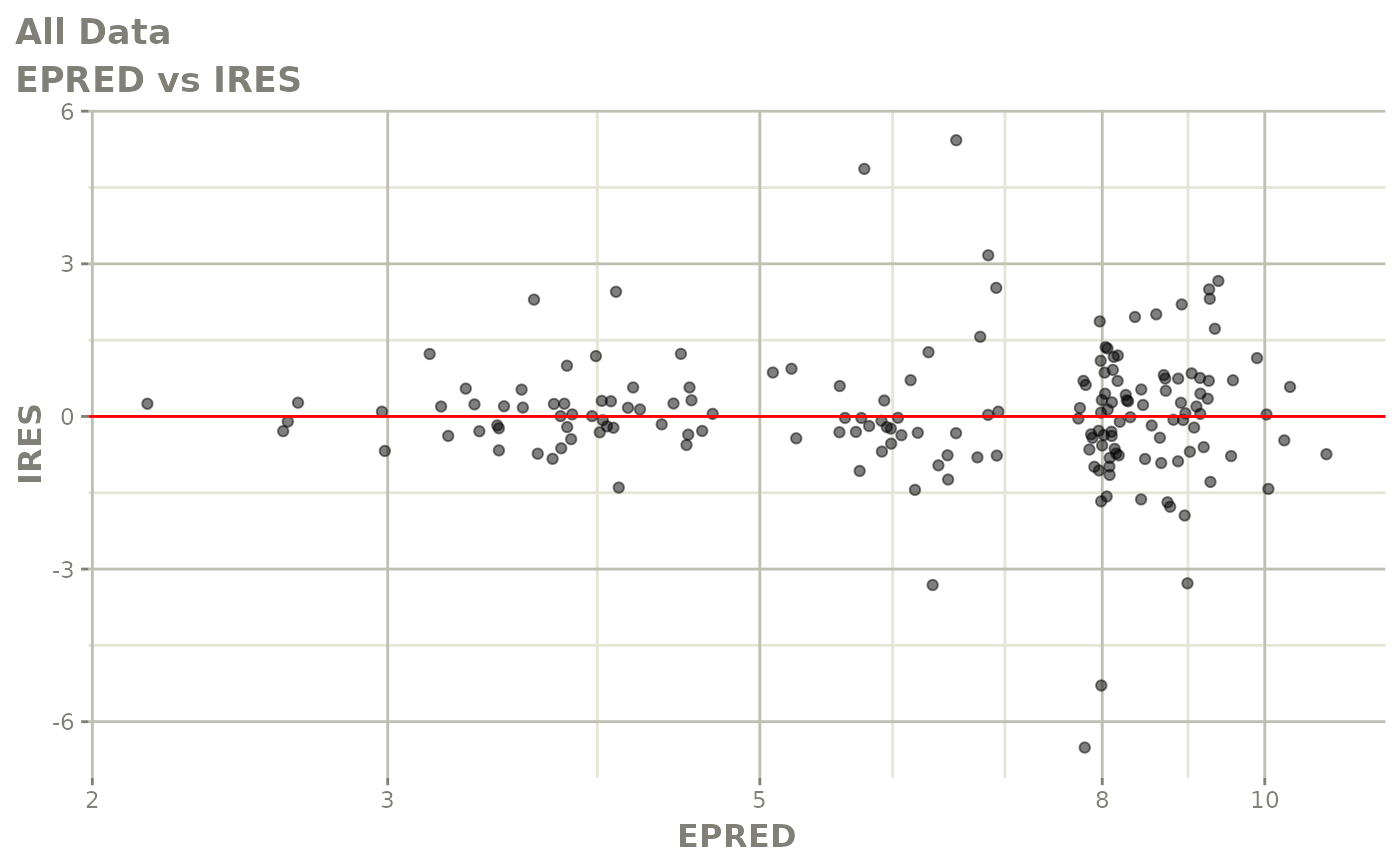
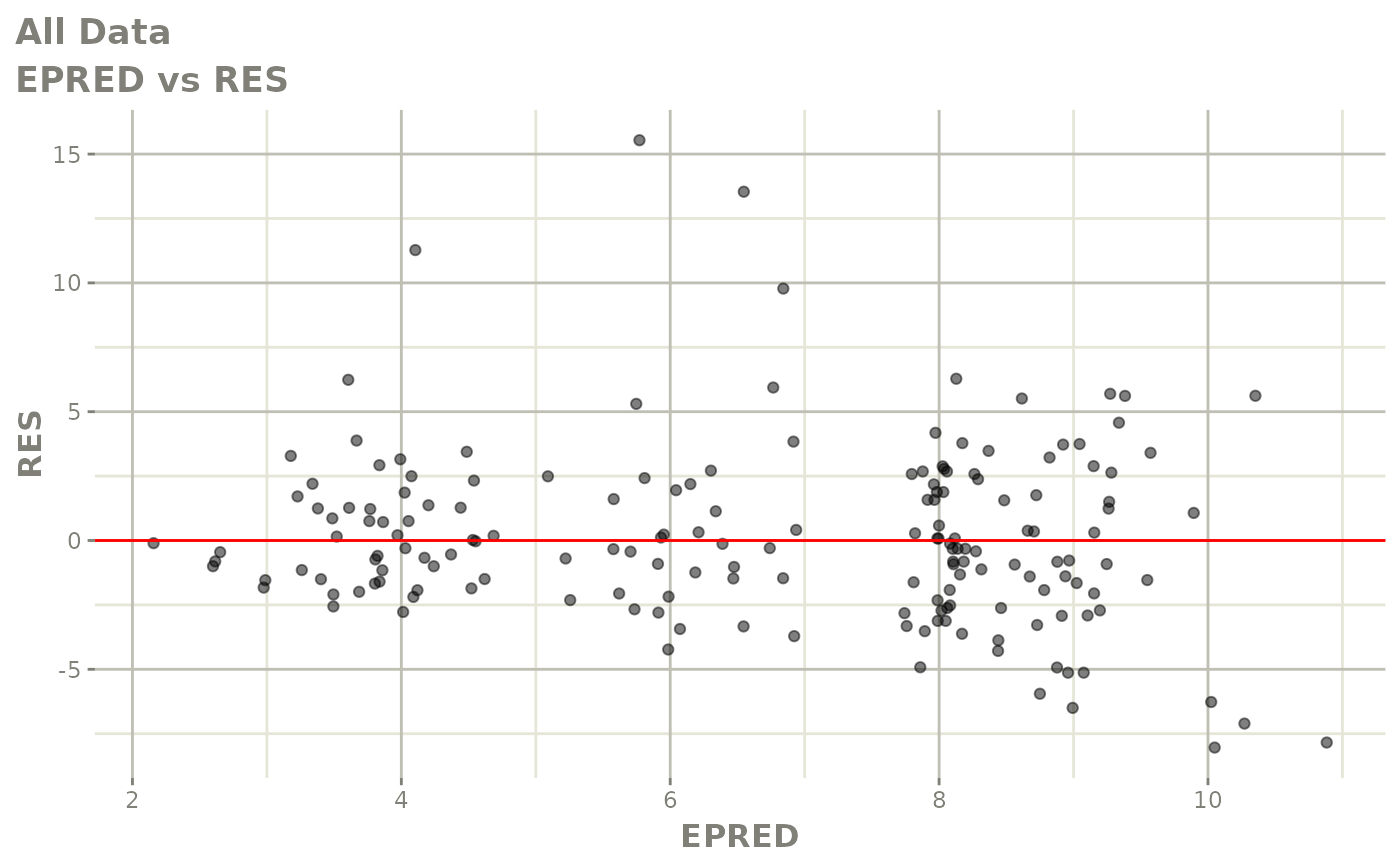
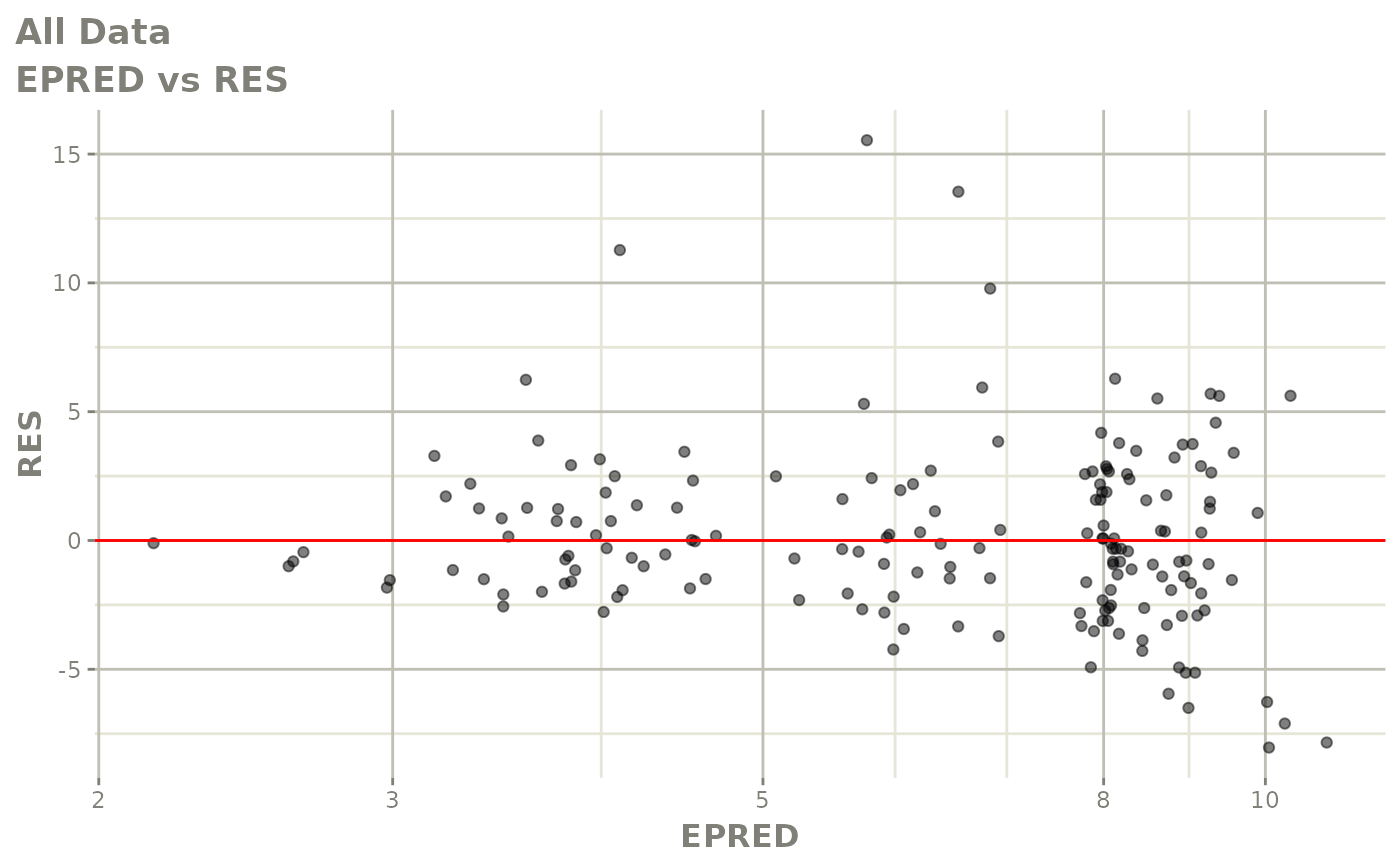
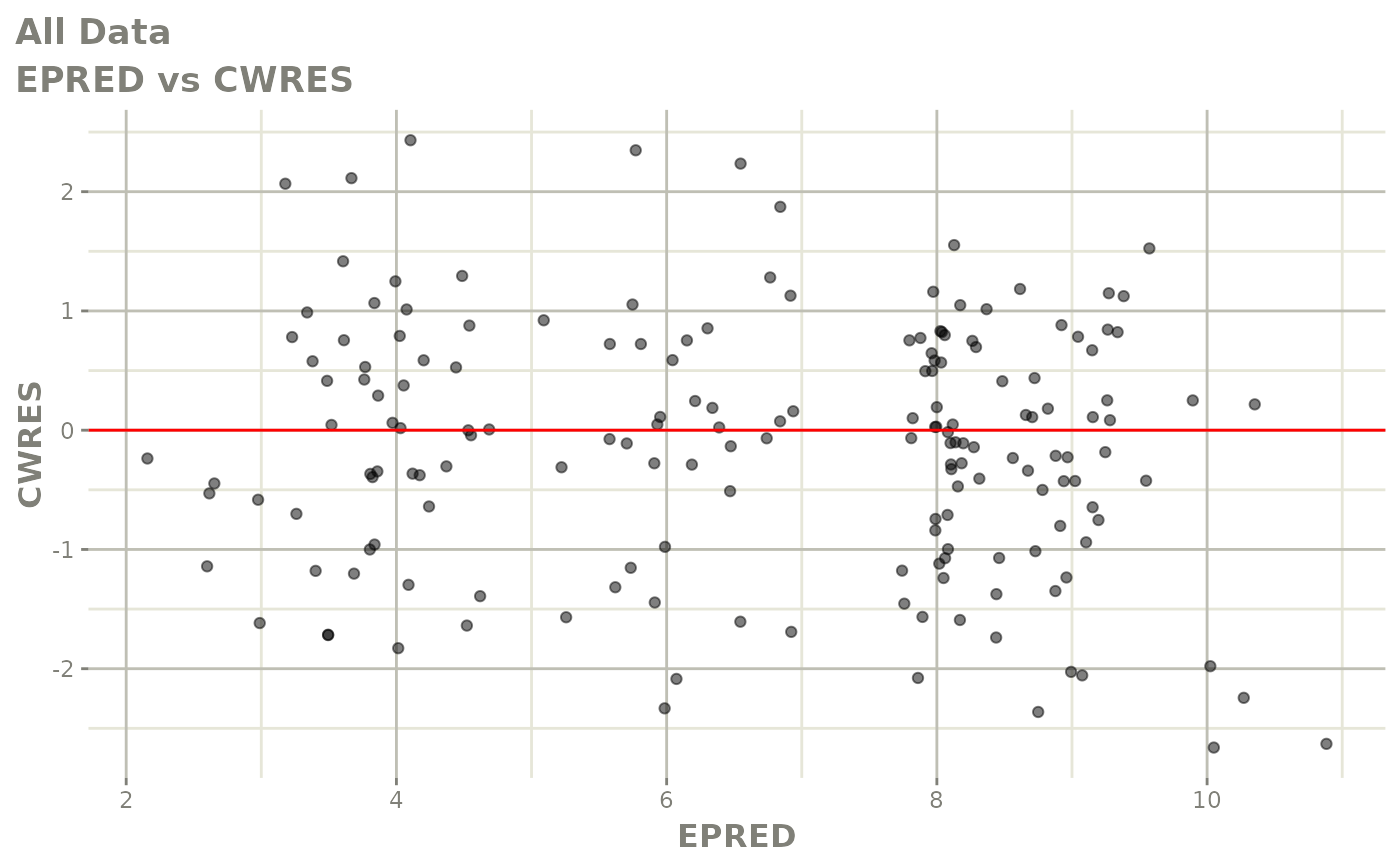
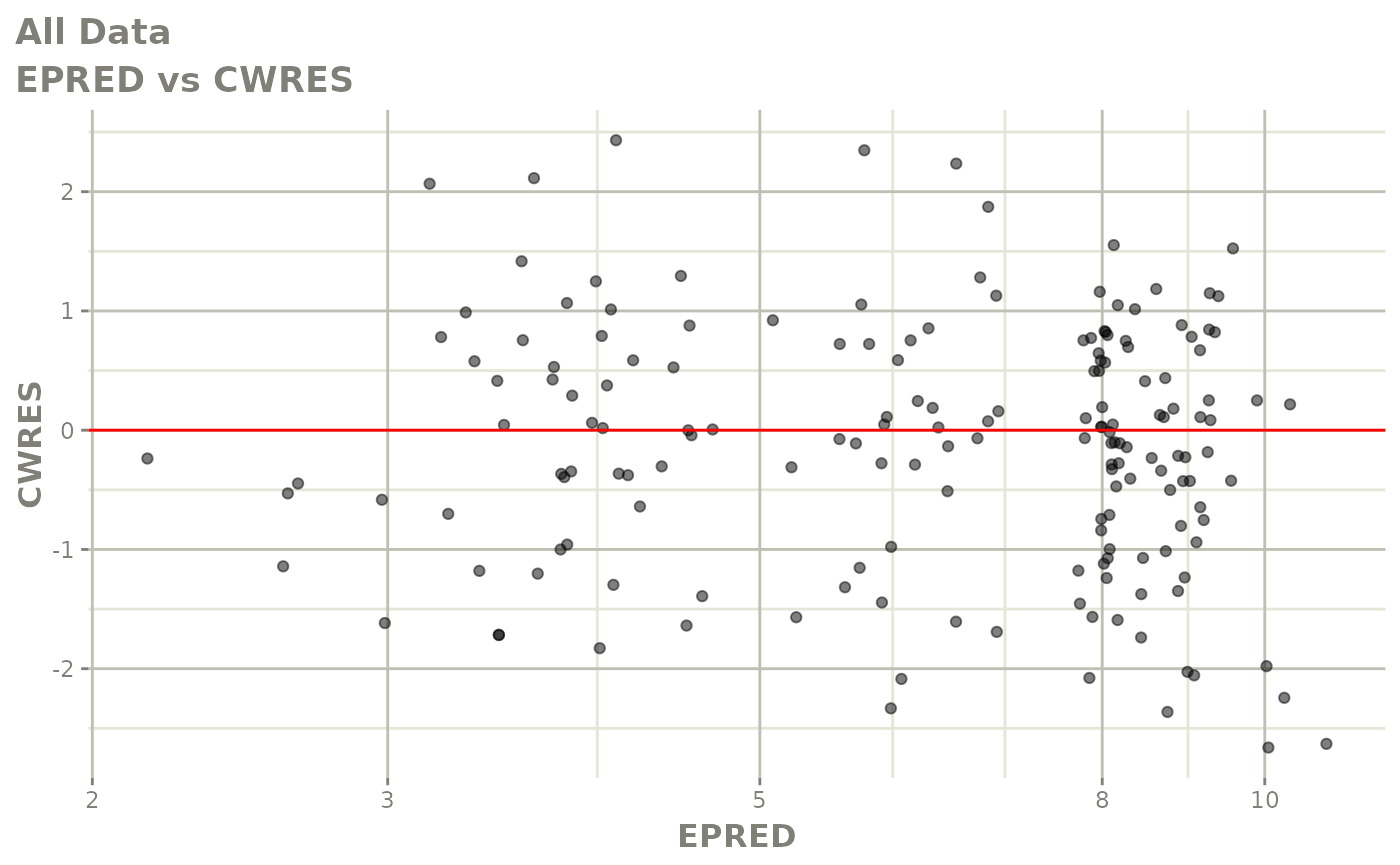
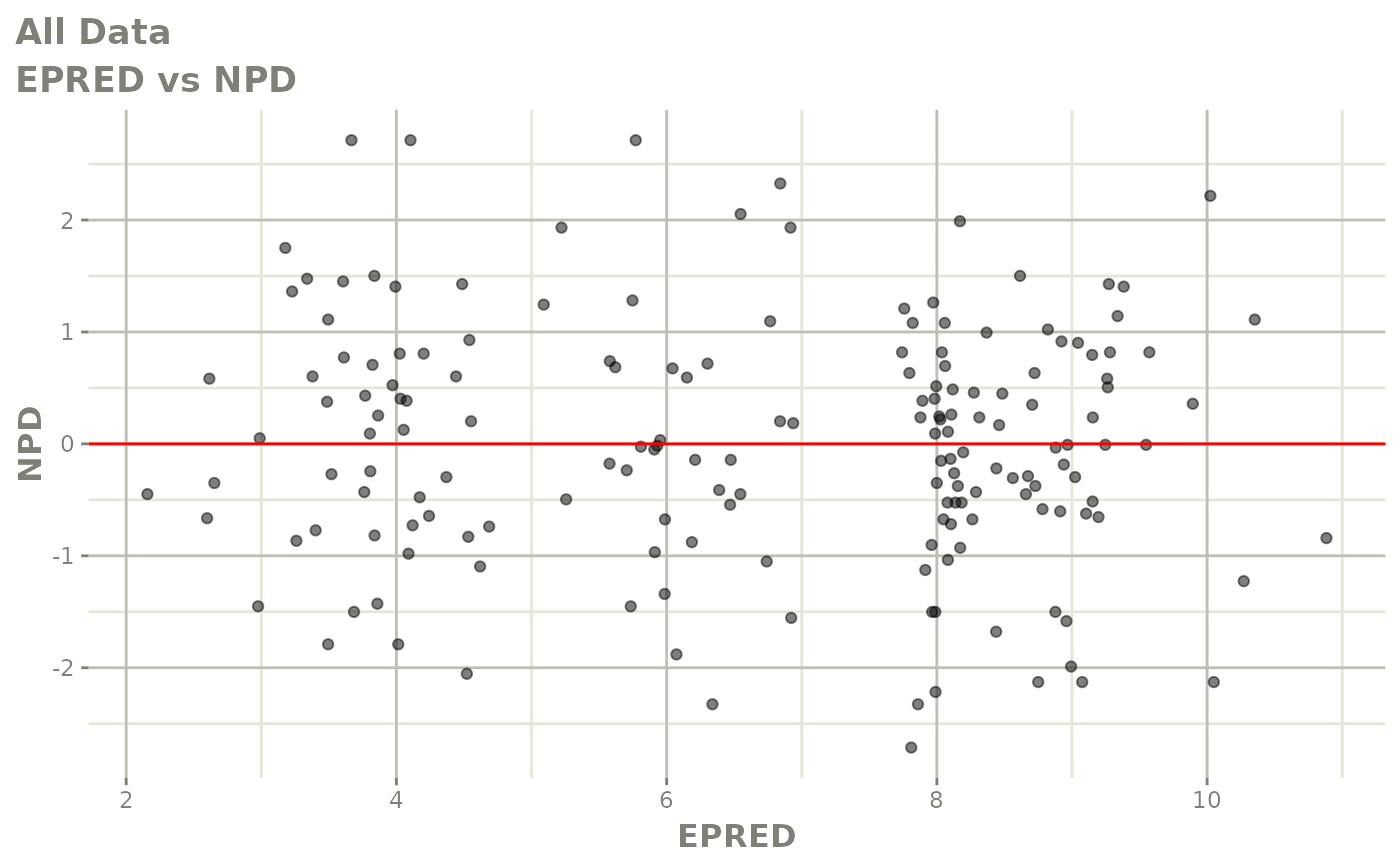
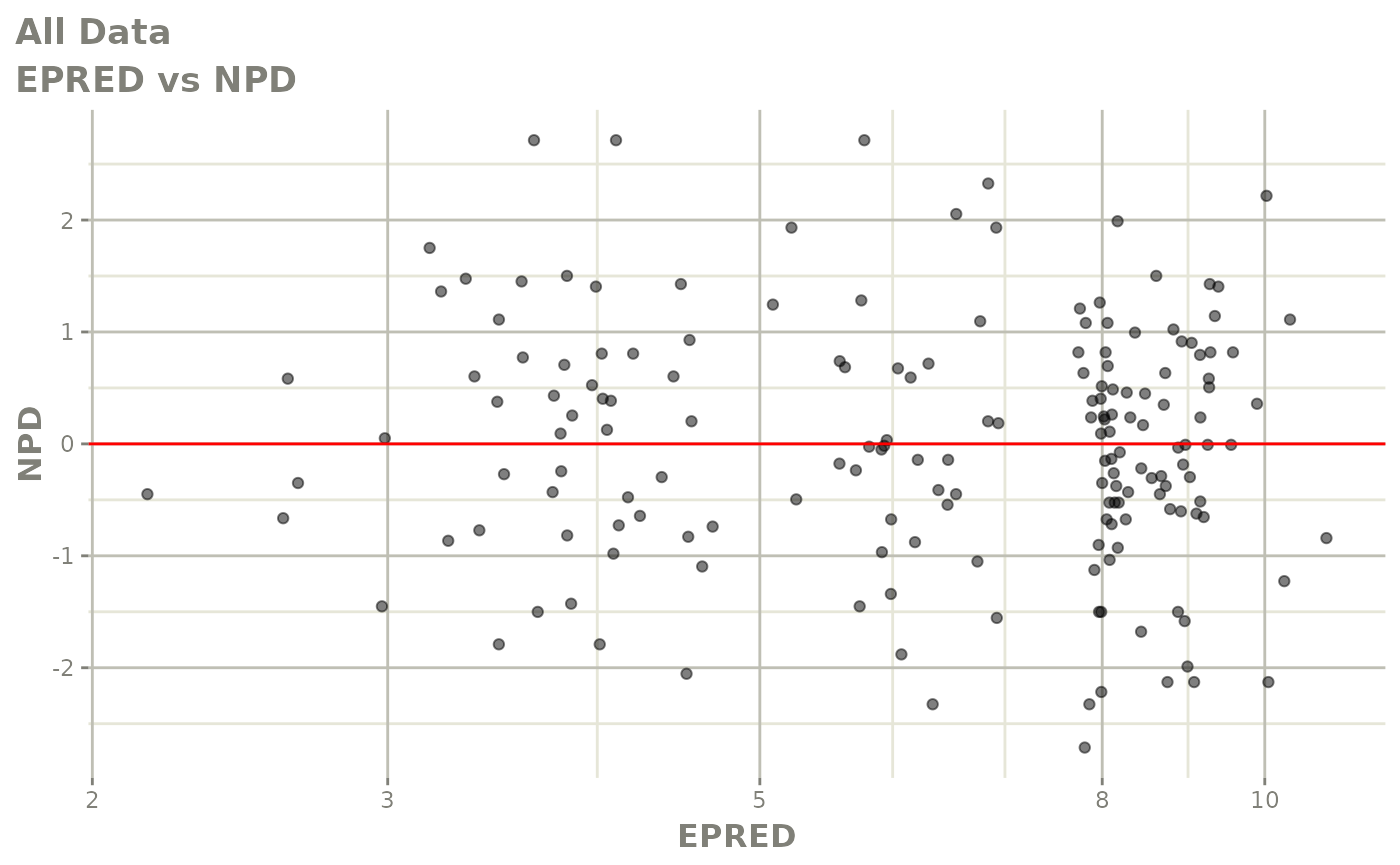
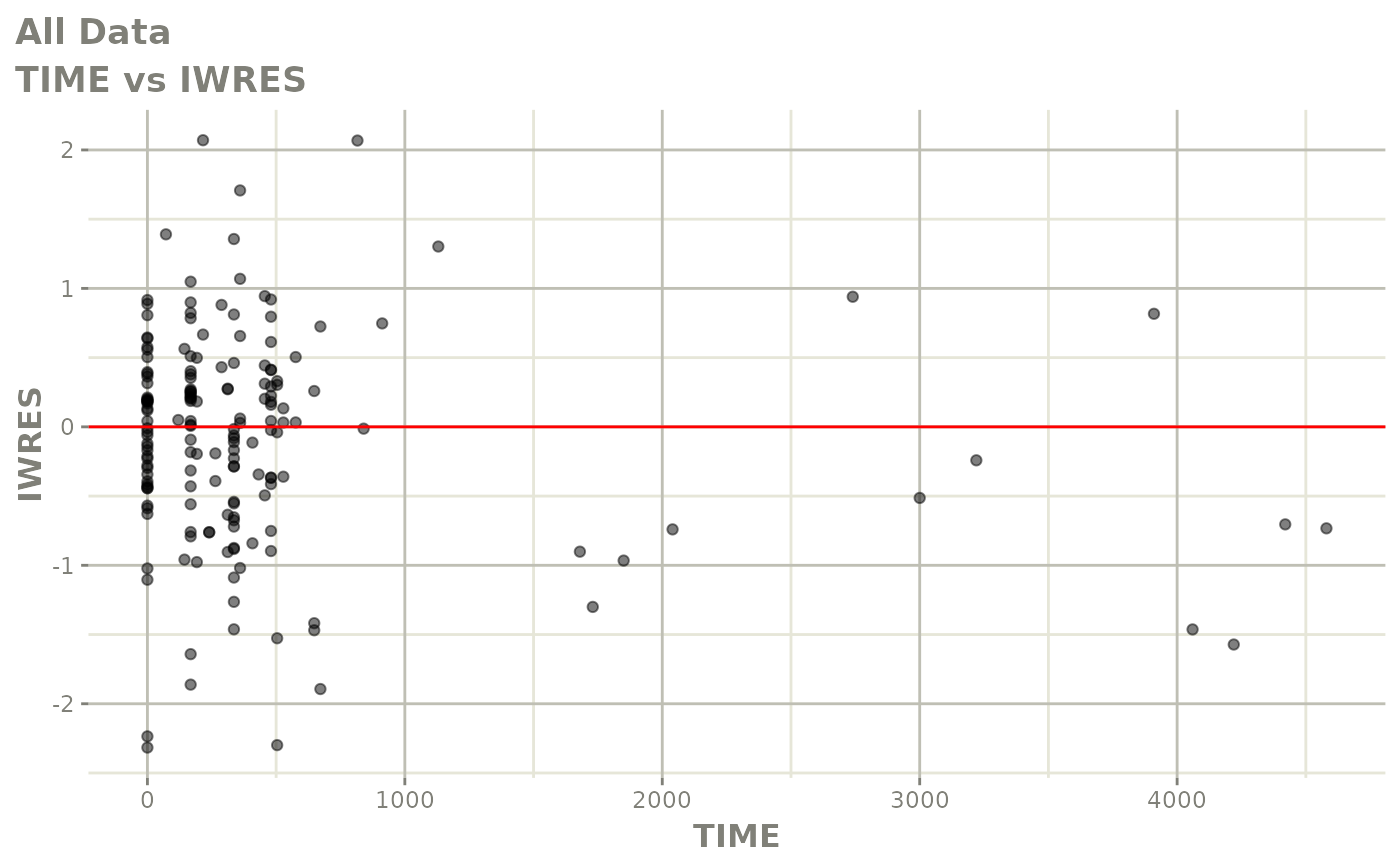
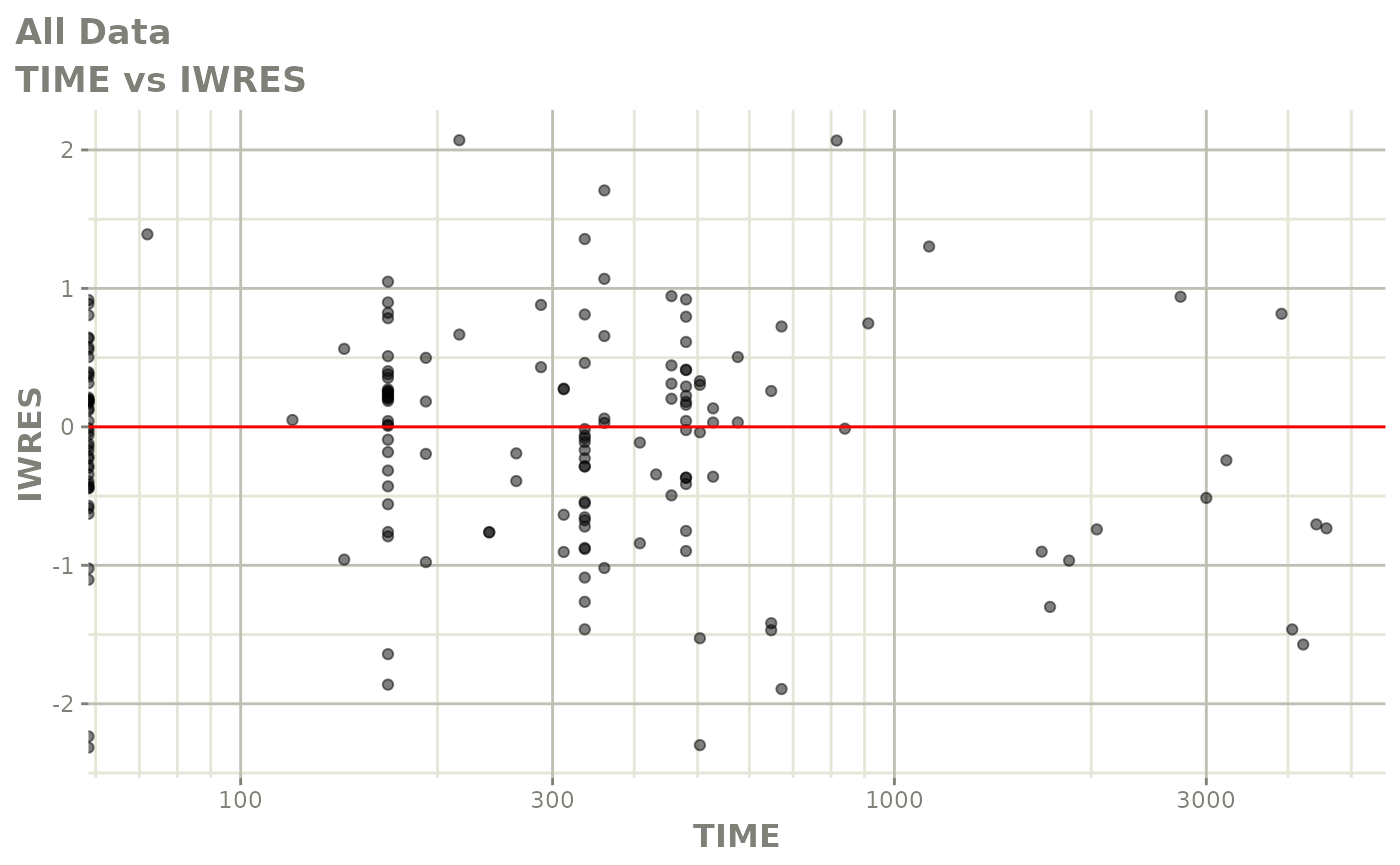
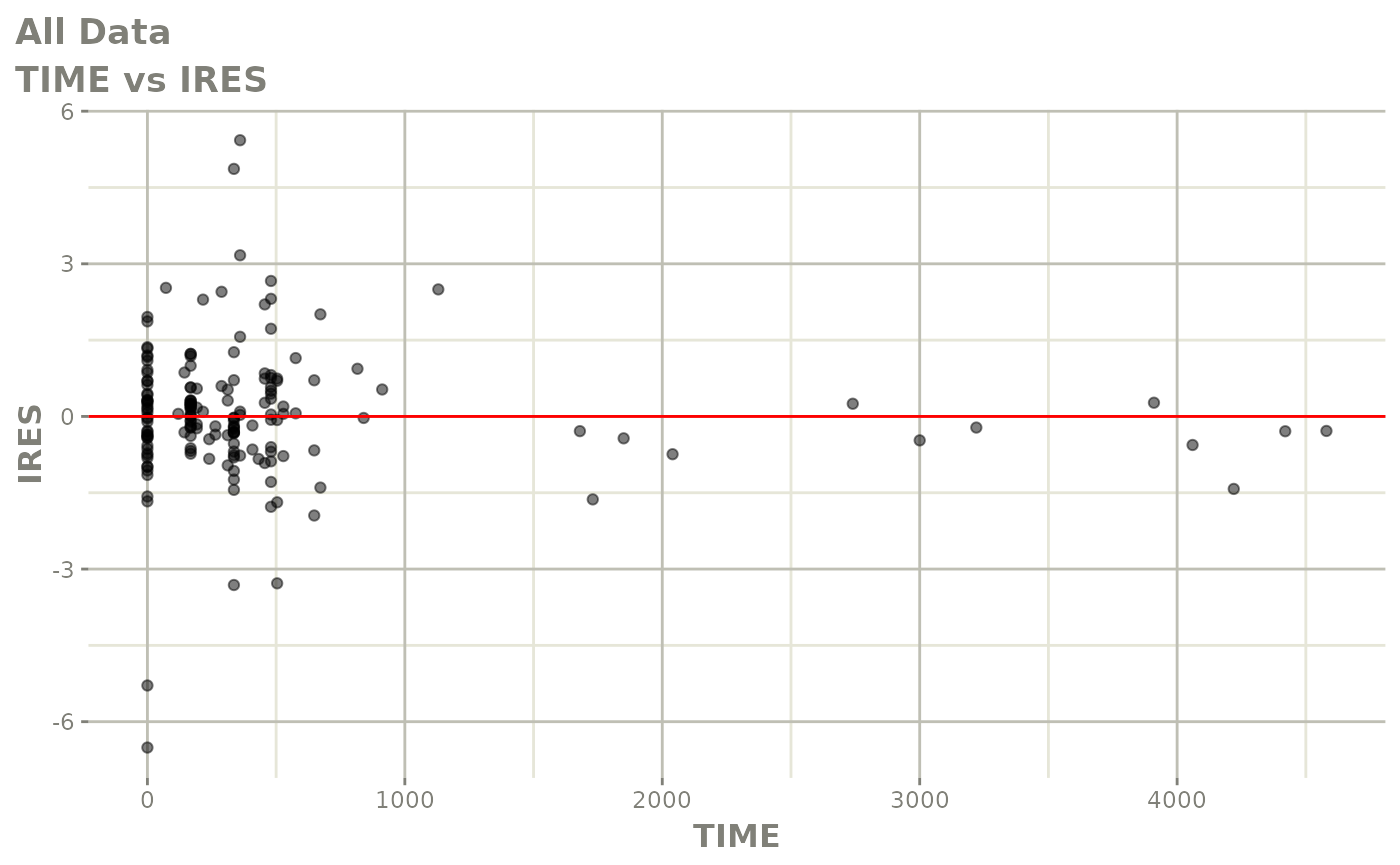
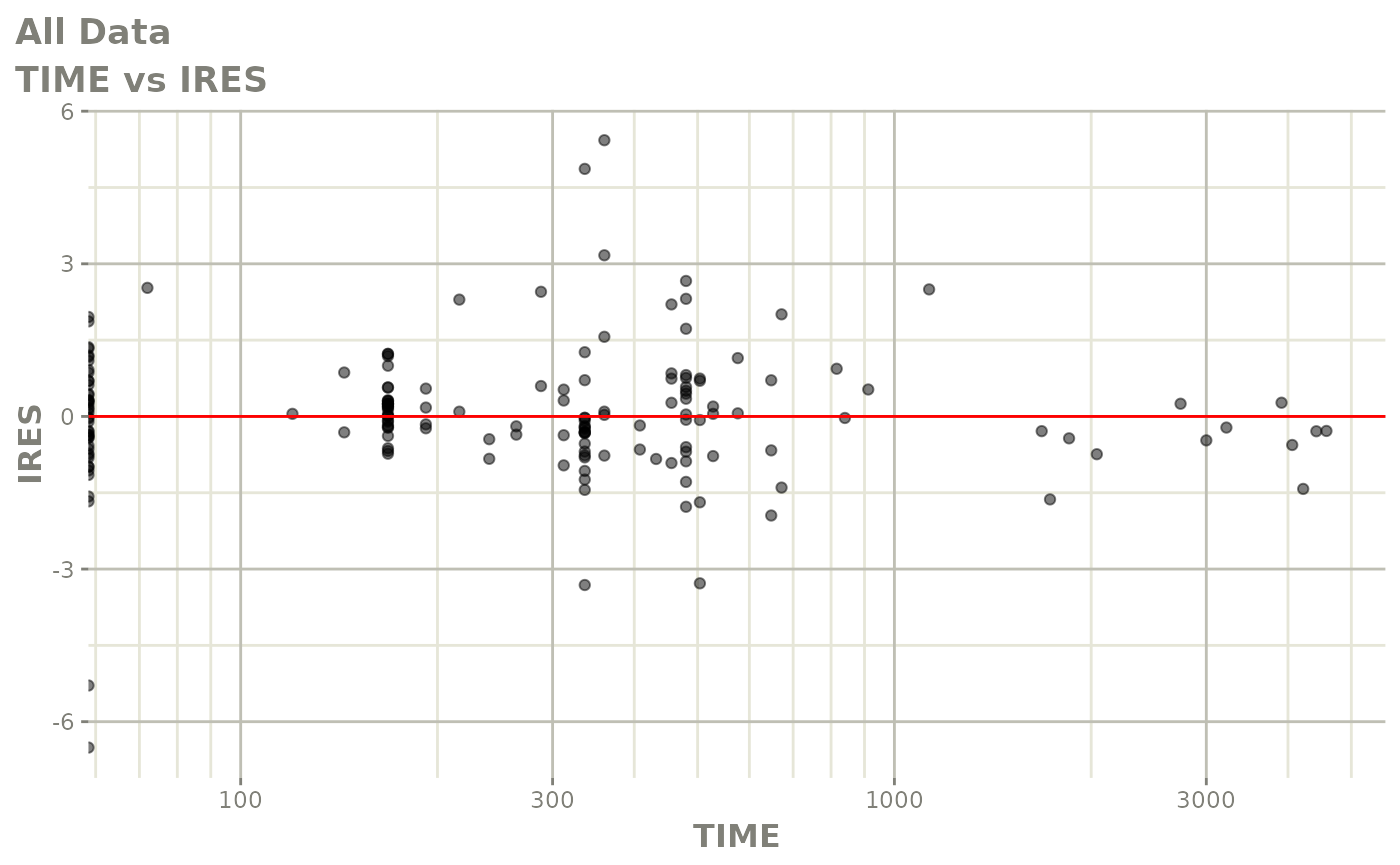
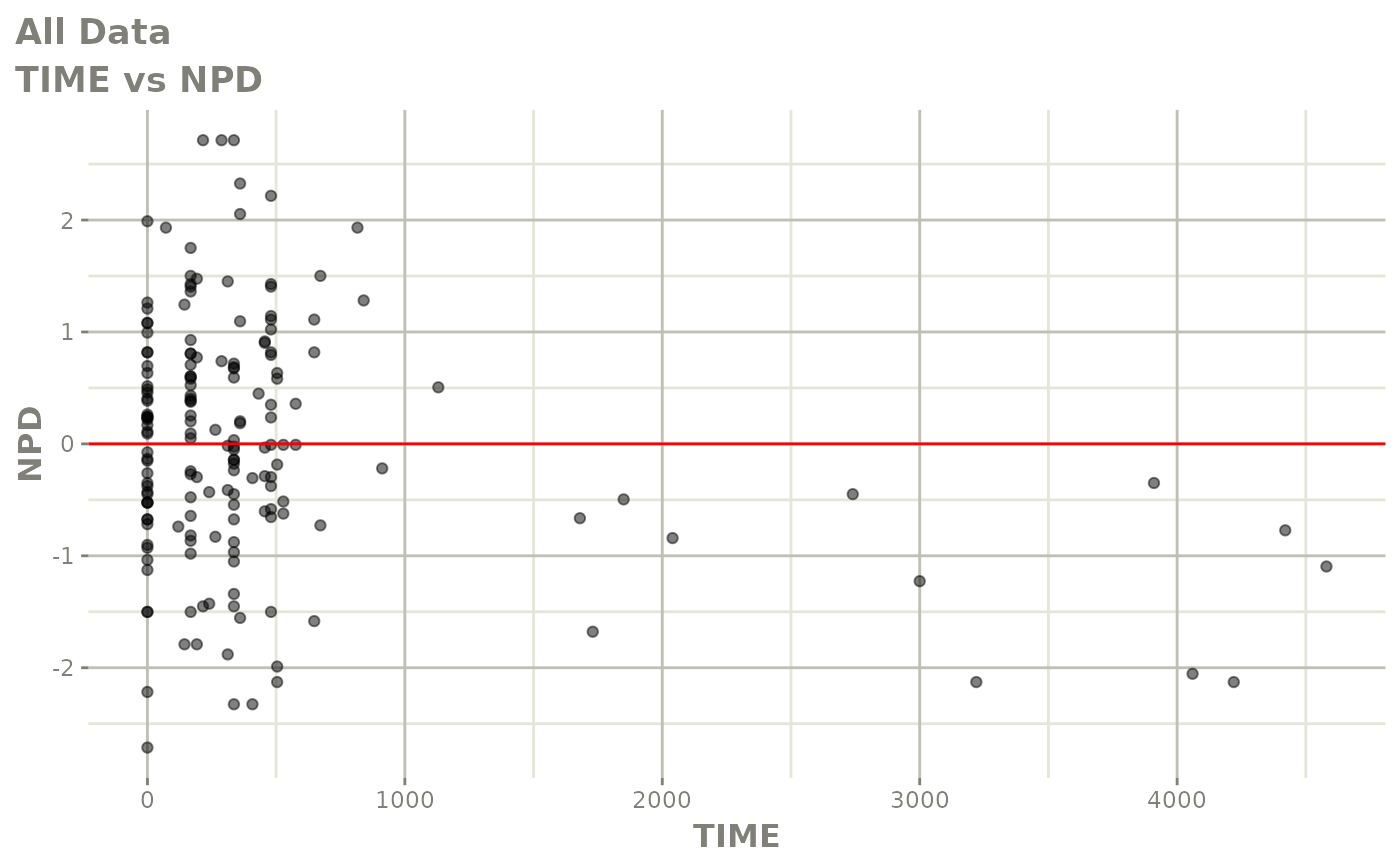
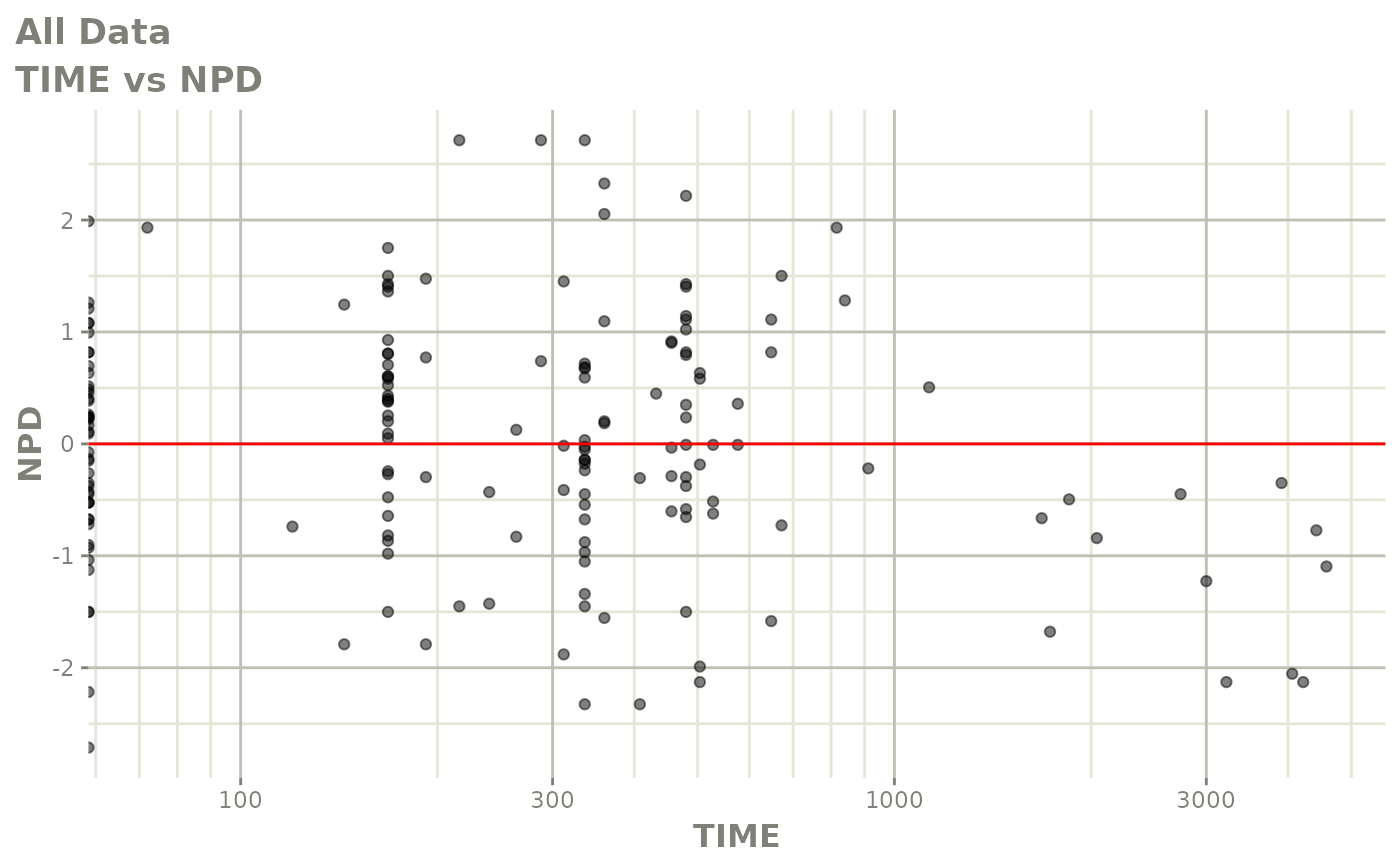
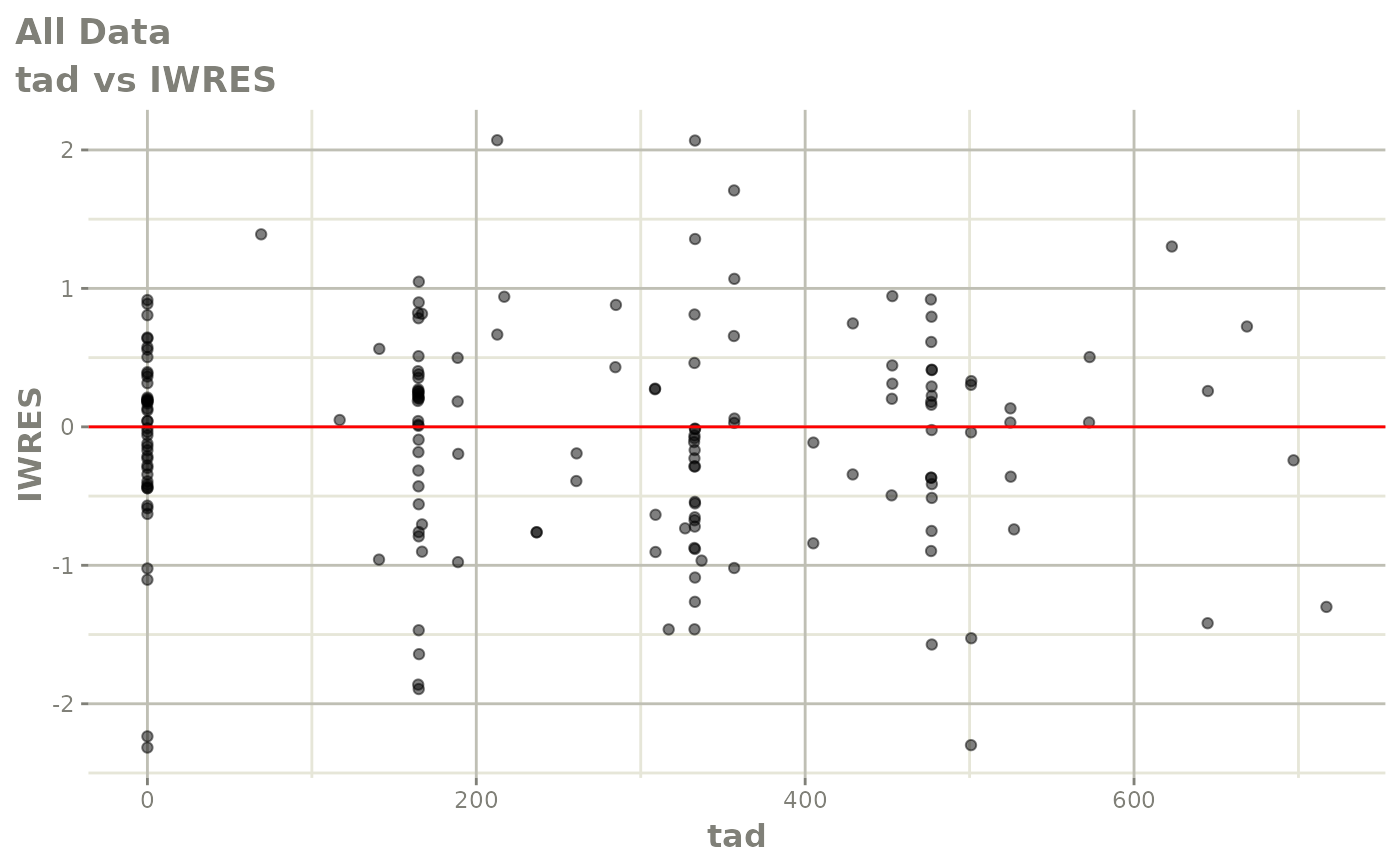
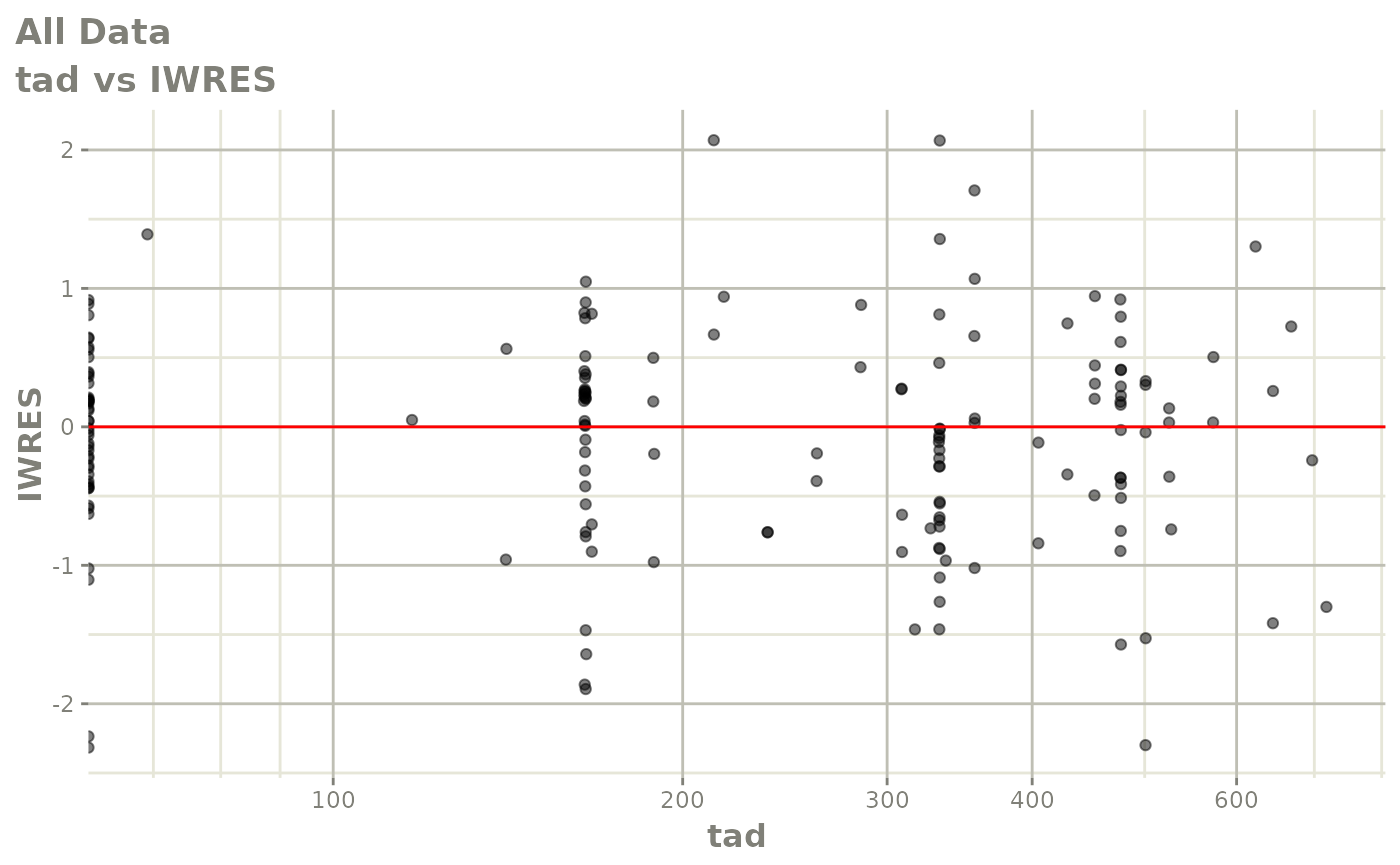
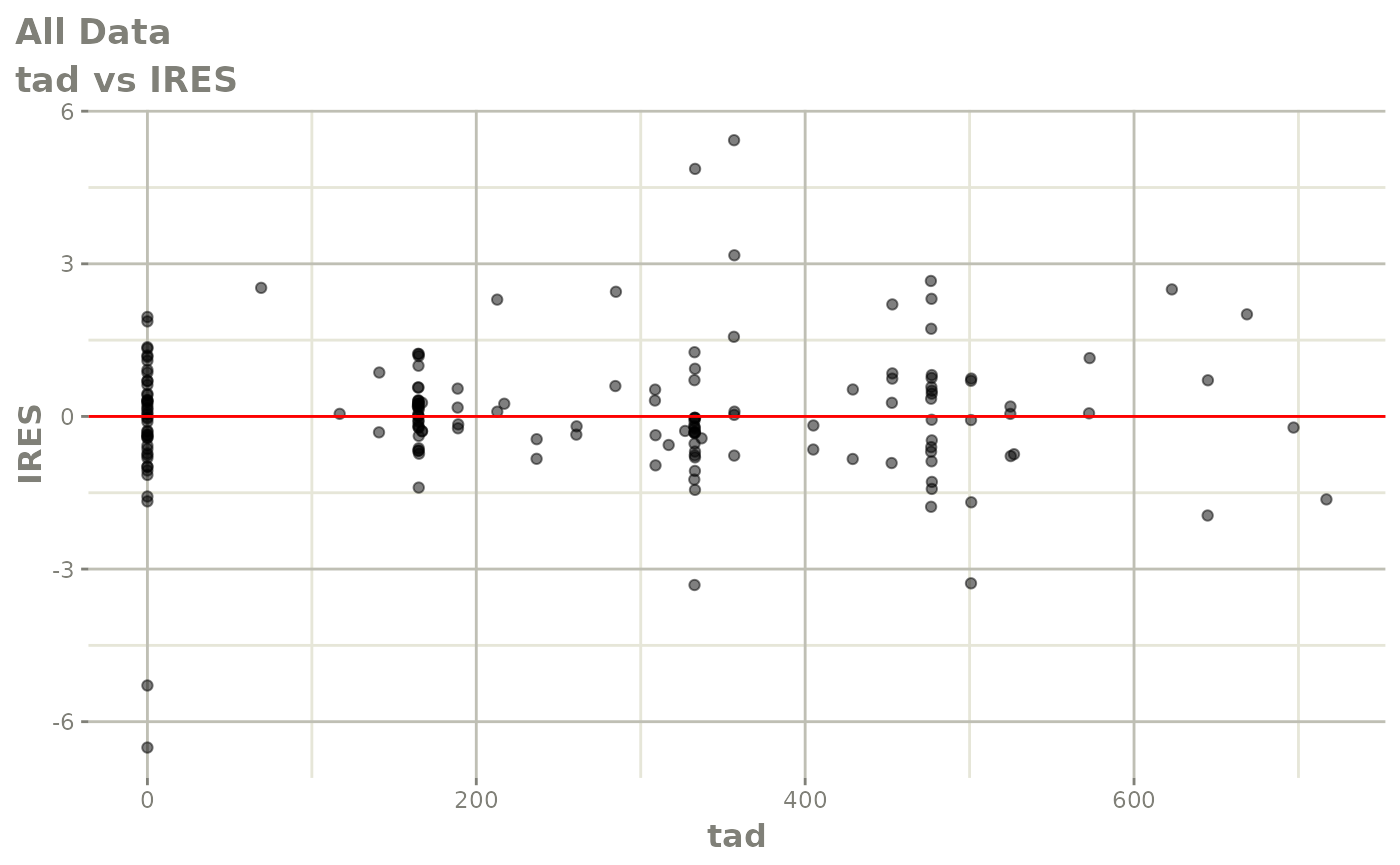
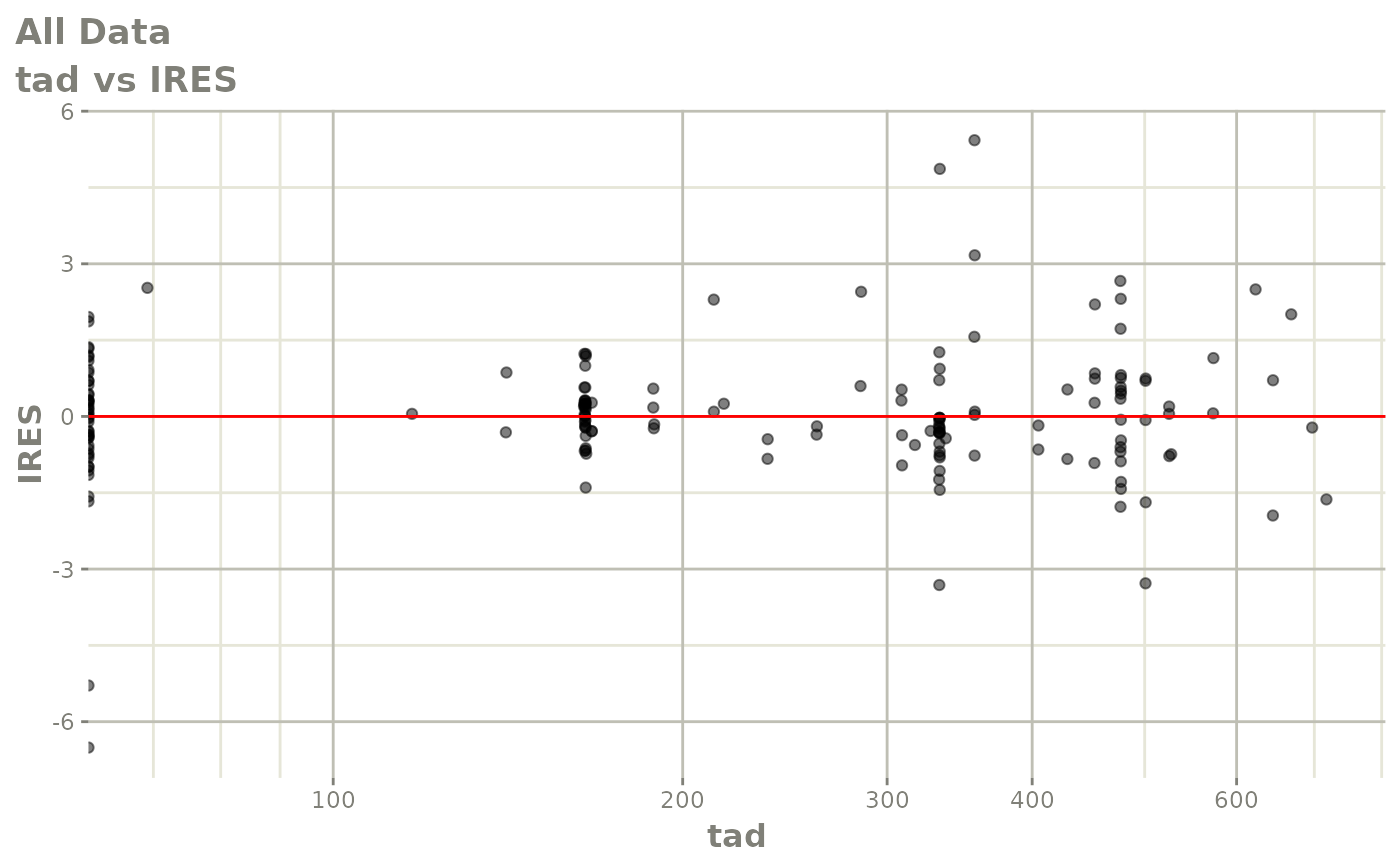
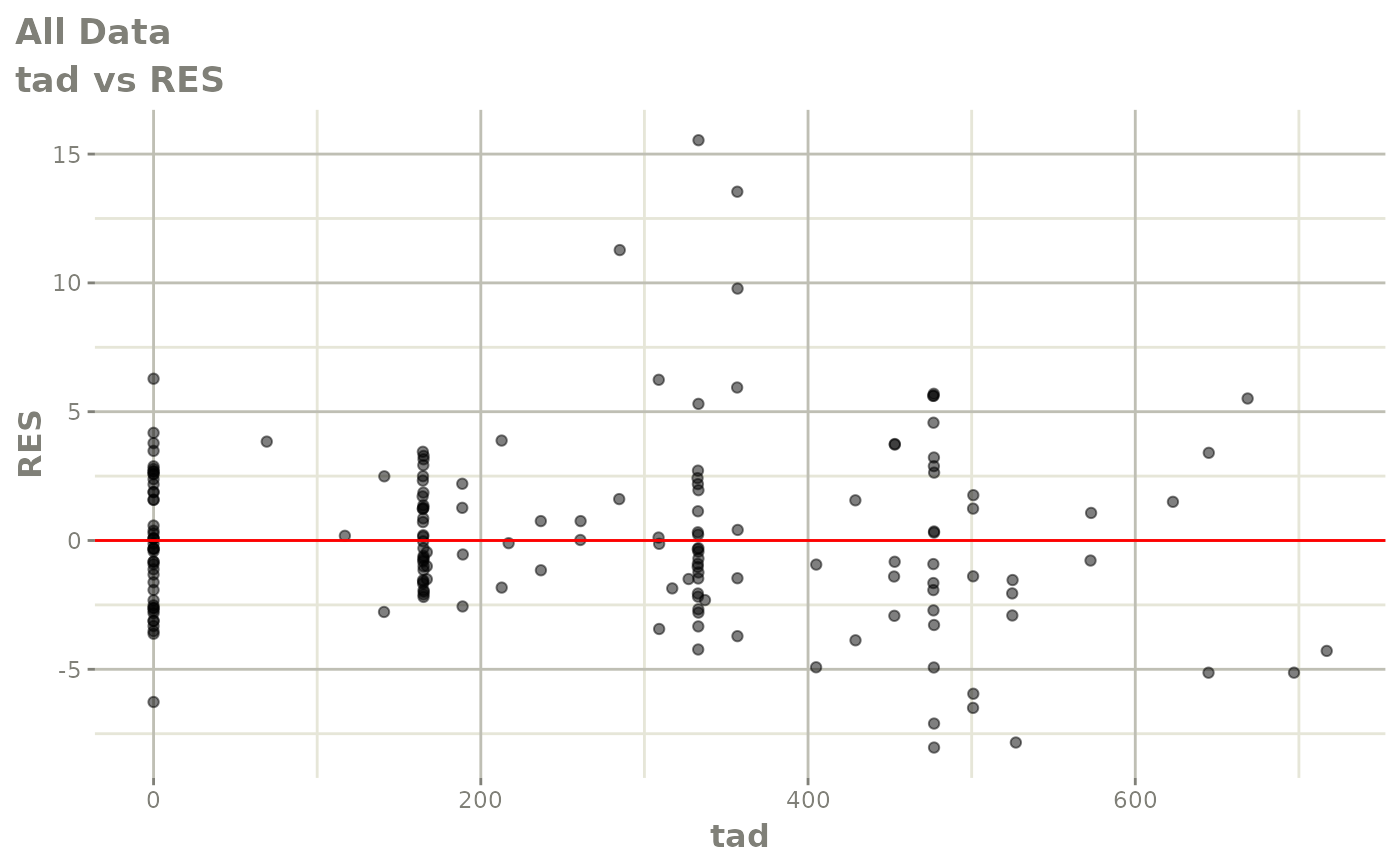
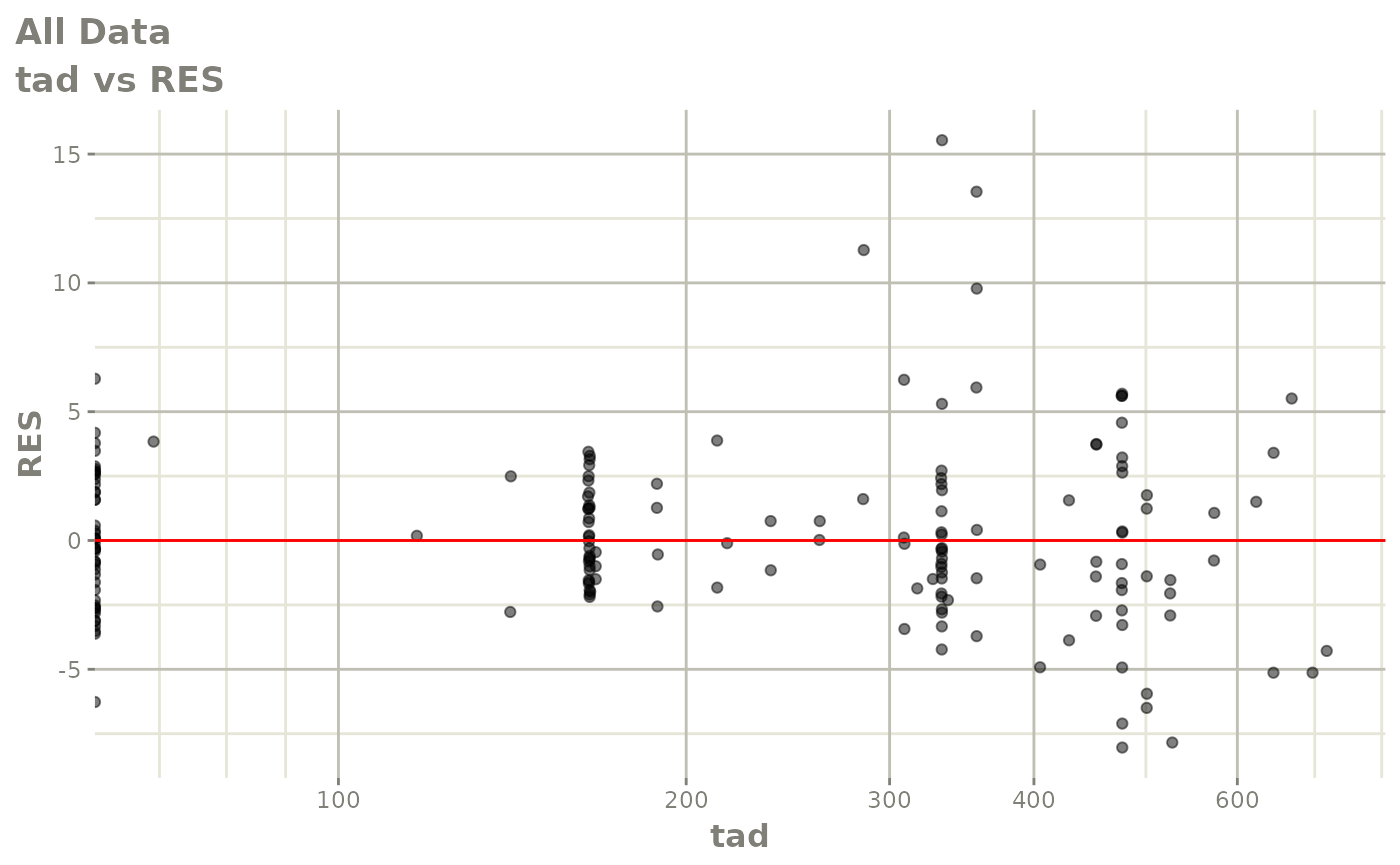
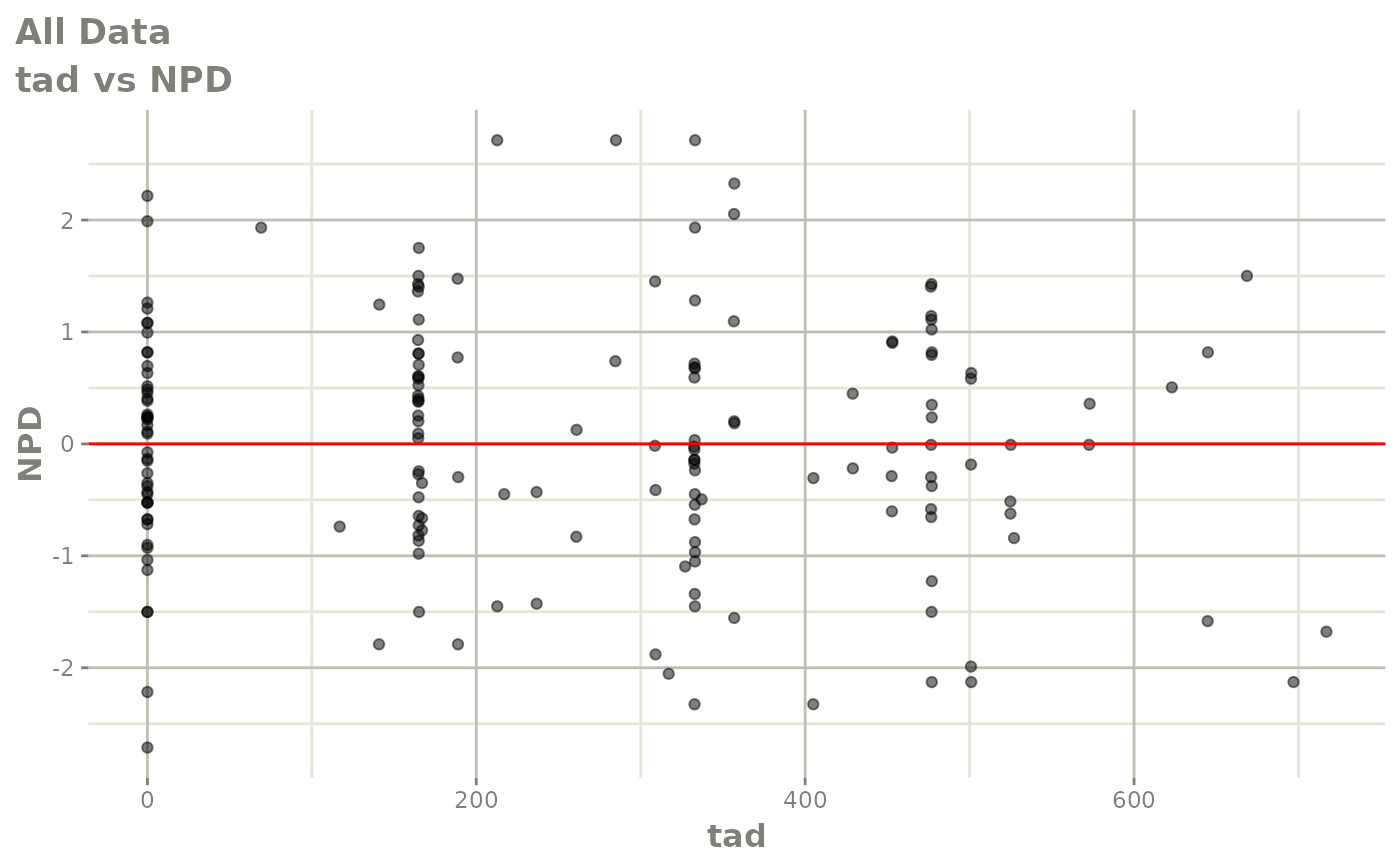
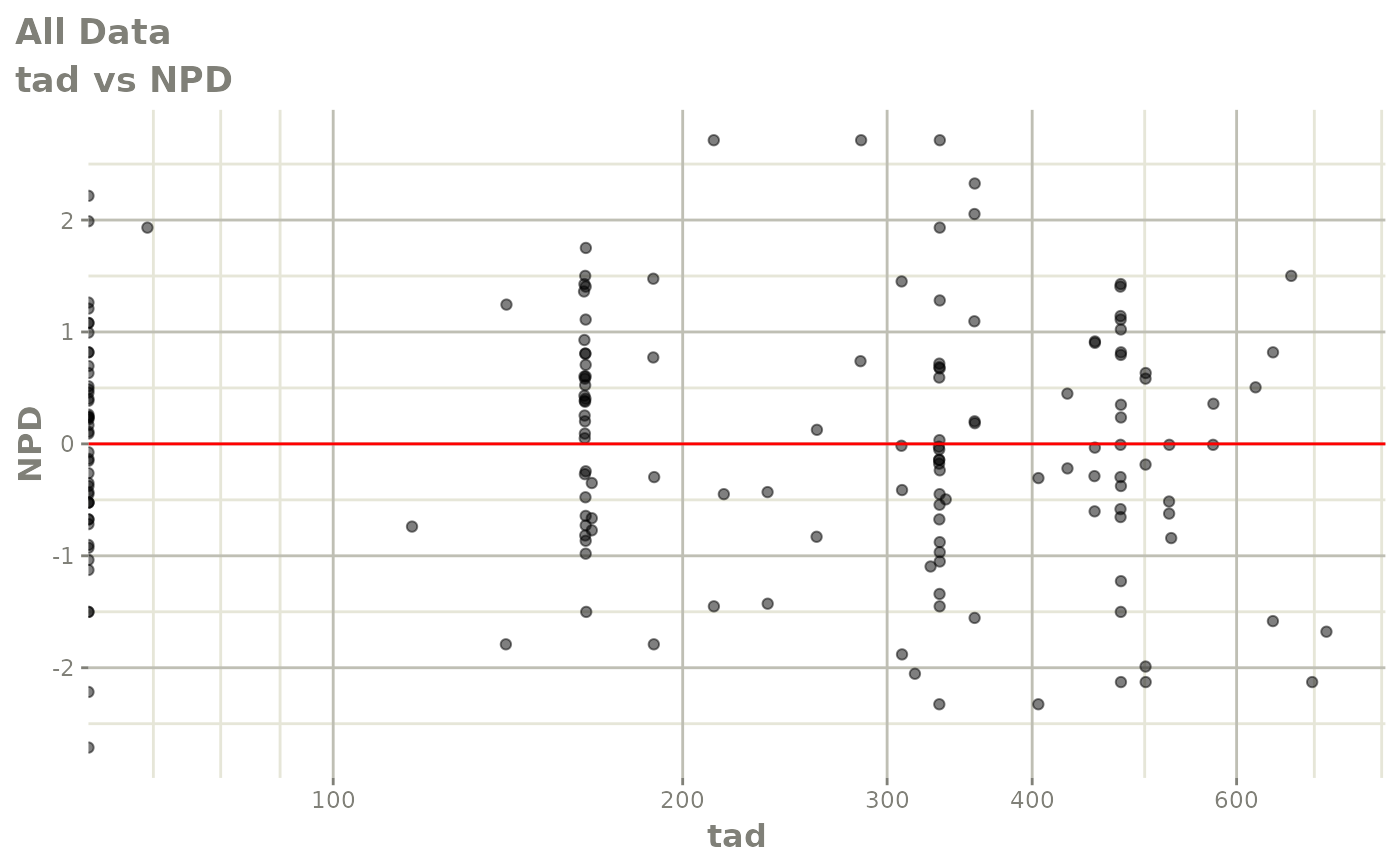
print(dv_vs_pred(xpdb) +
ylab("Observed Neutrophil Count (10^9/L)") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))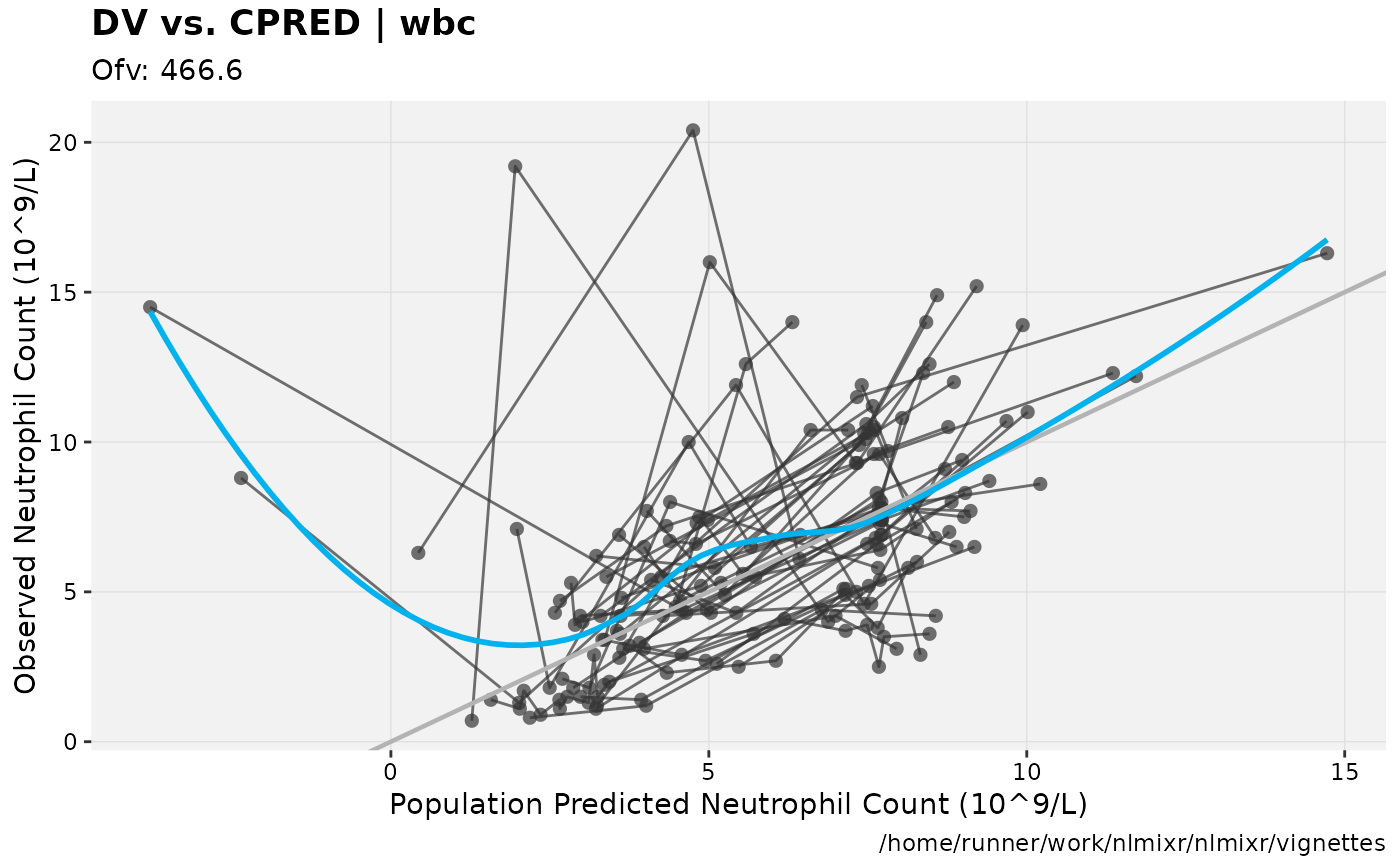
print(dv_vs_ipred(xpdb) +
ylab("Observed Neutrophil Count (10^9/L)") +
xlab("Individual Predicted Neutrophil Count (10^9/L)"))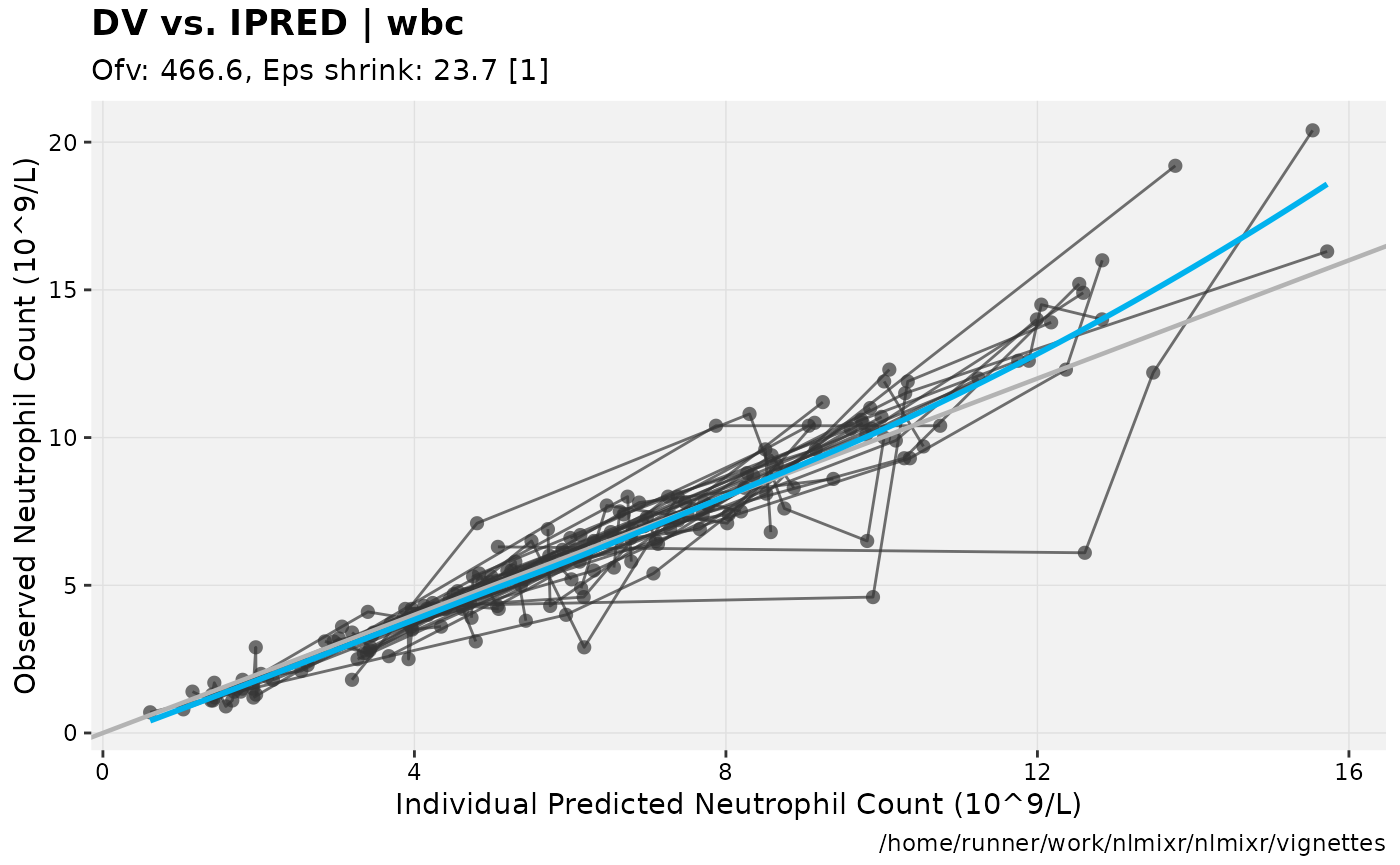
print(res_vs_pred(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))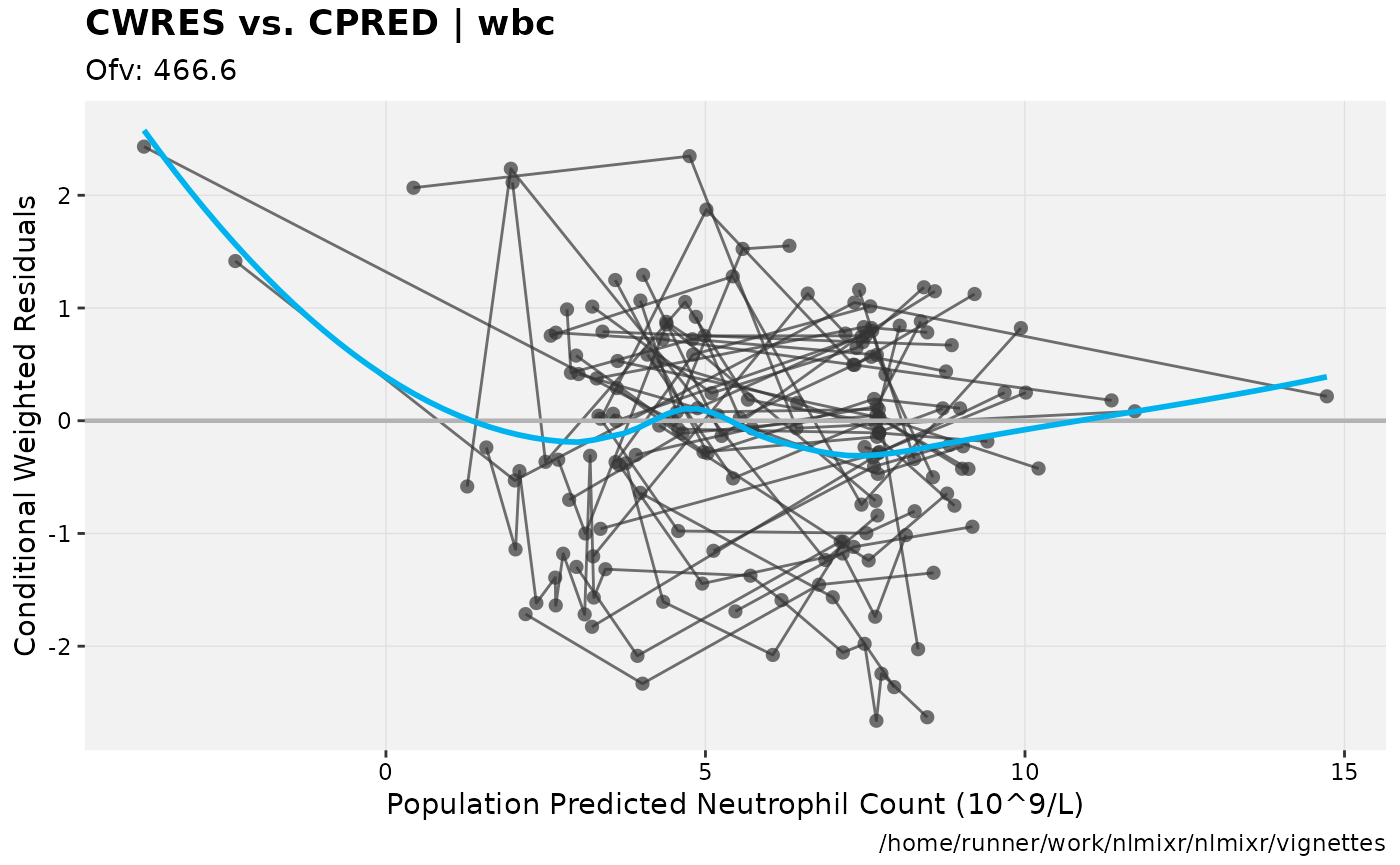
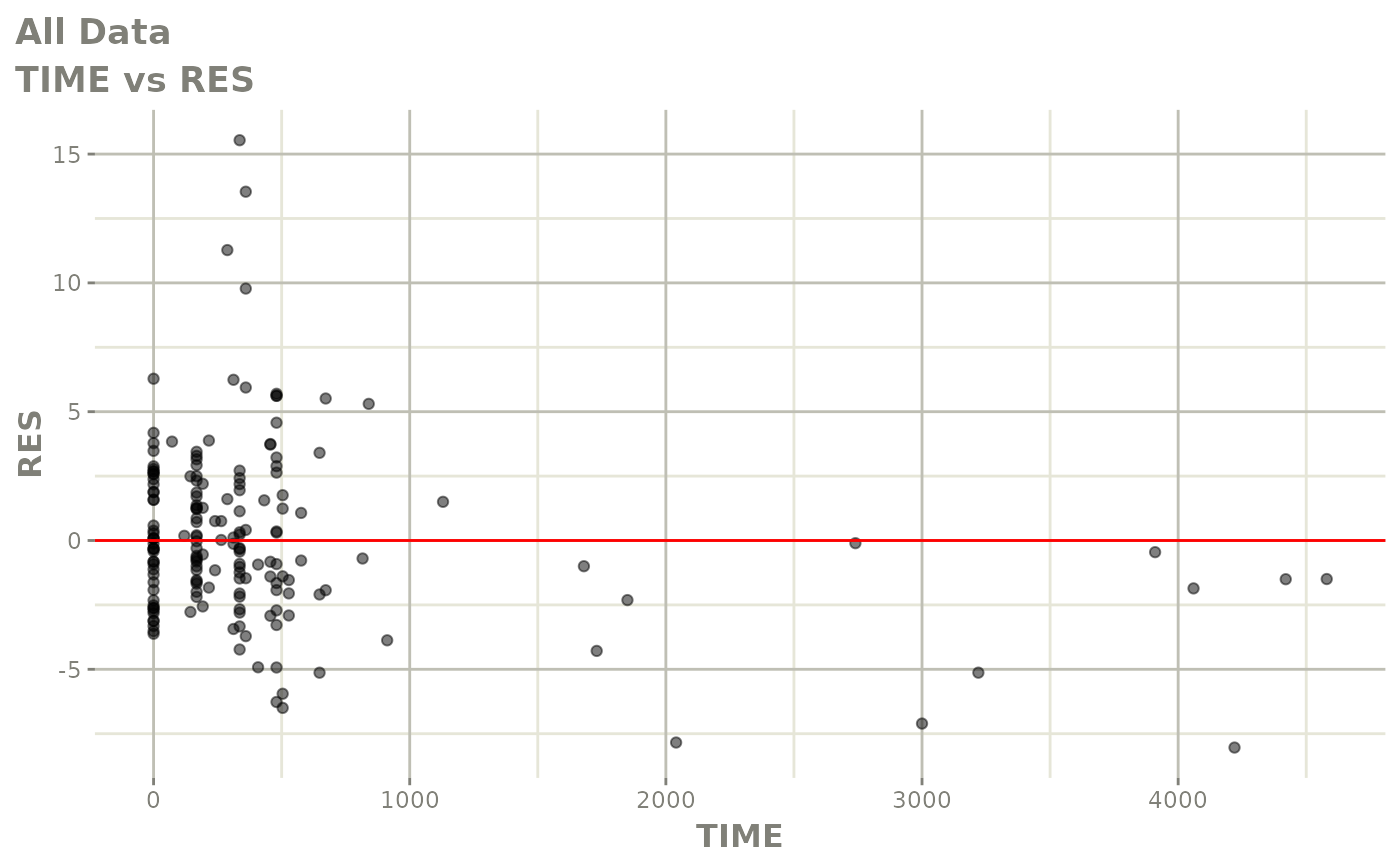
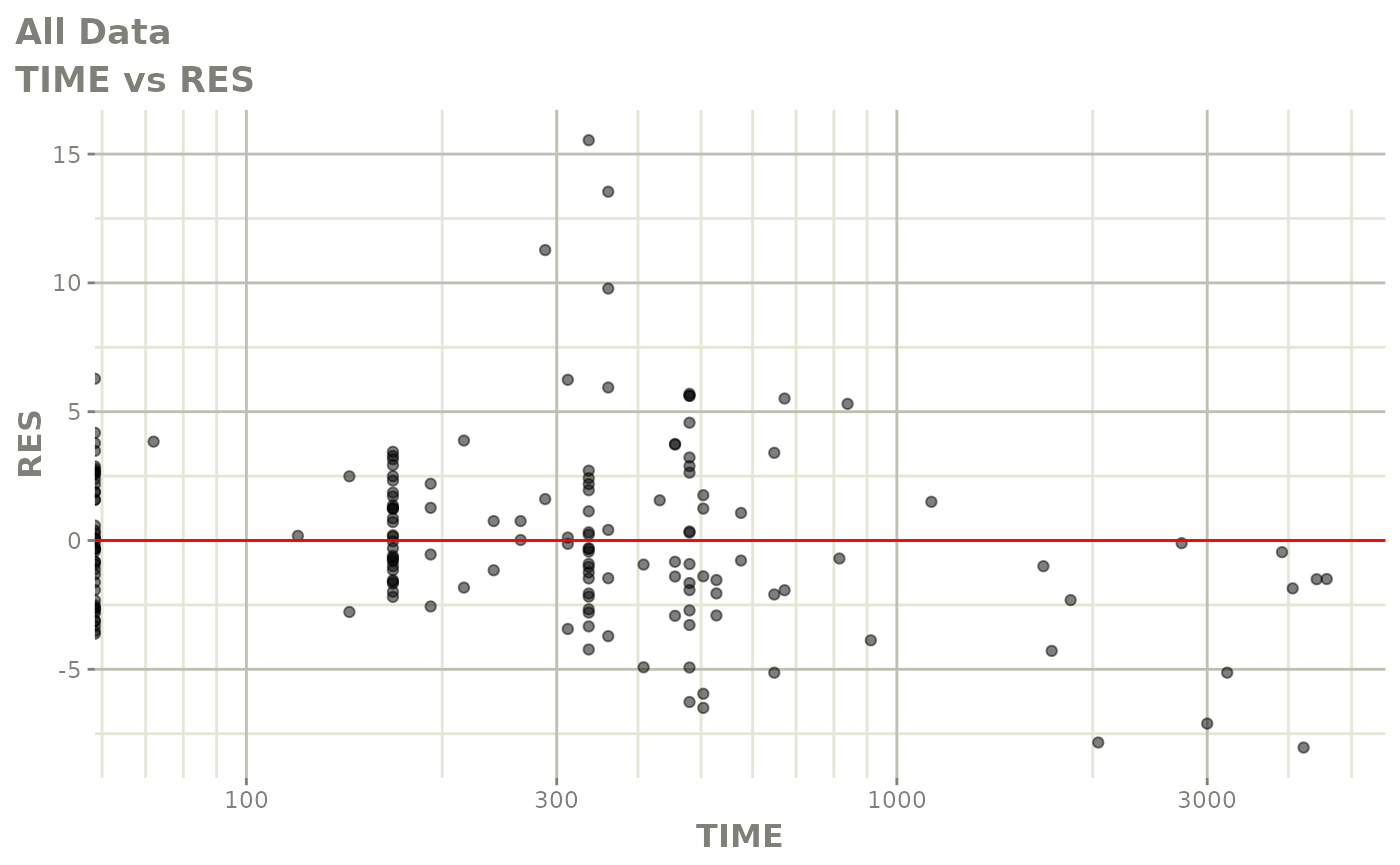
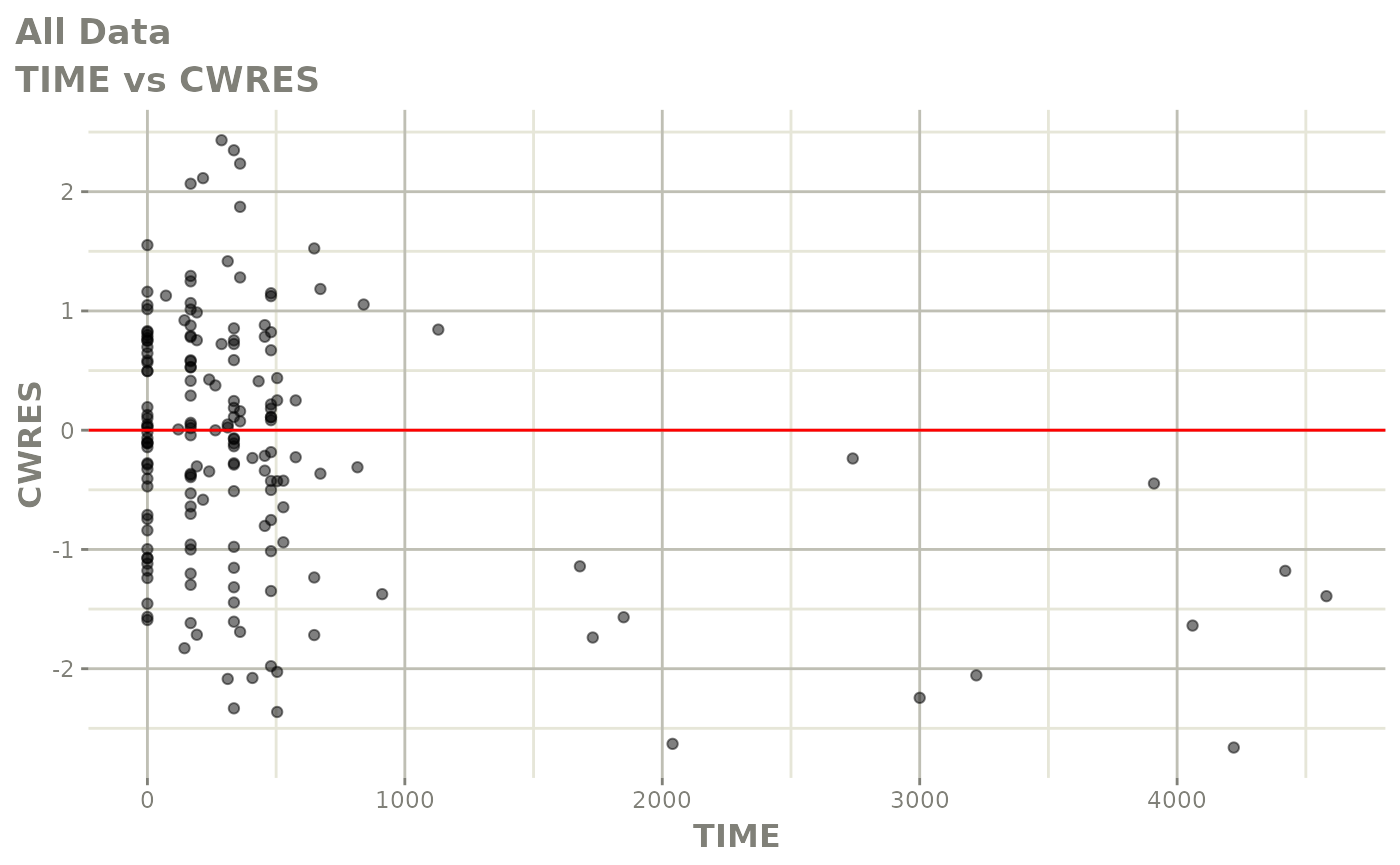
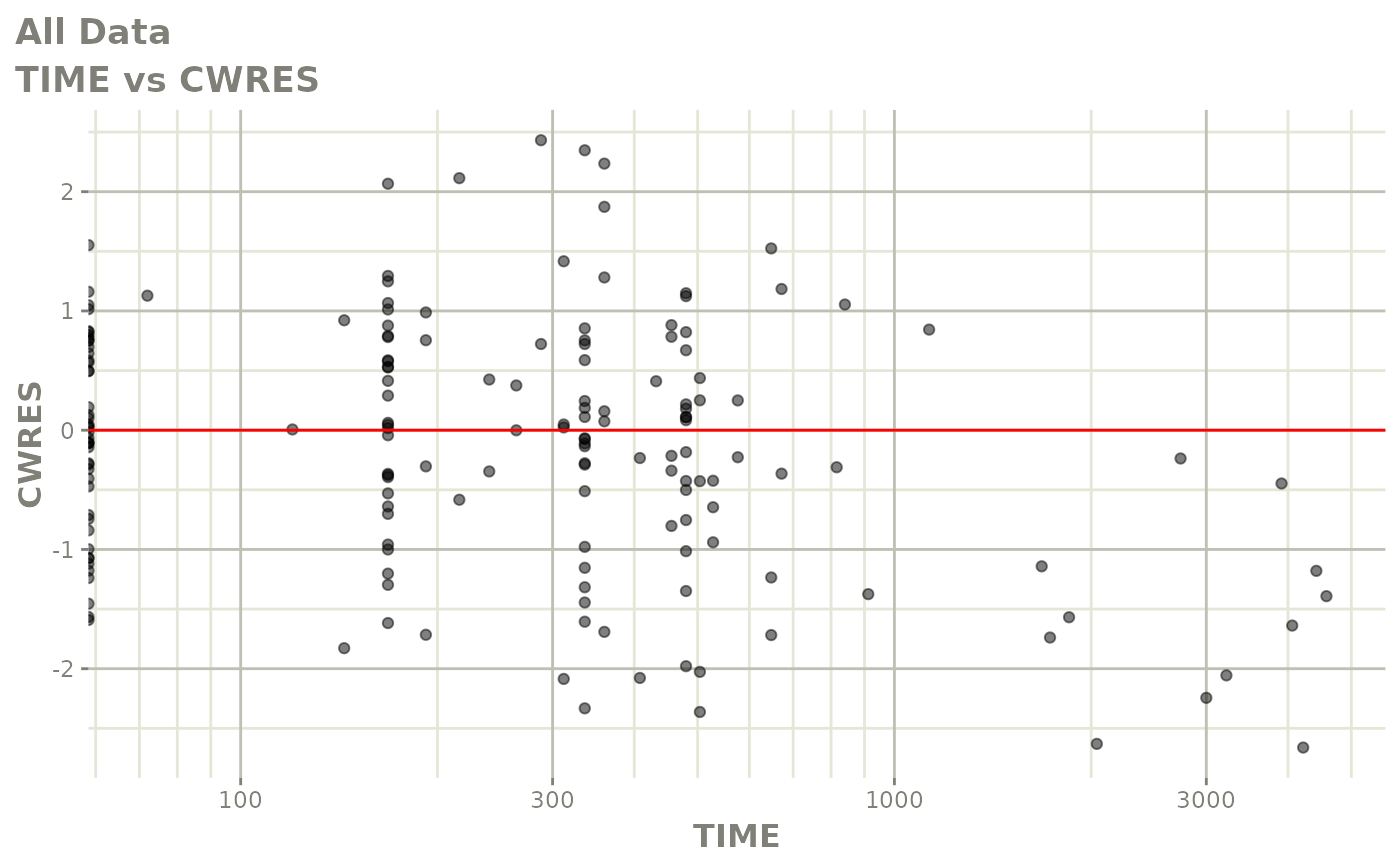
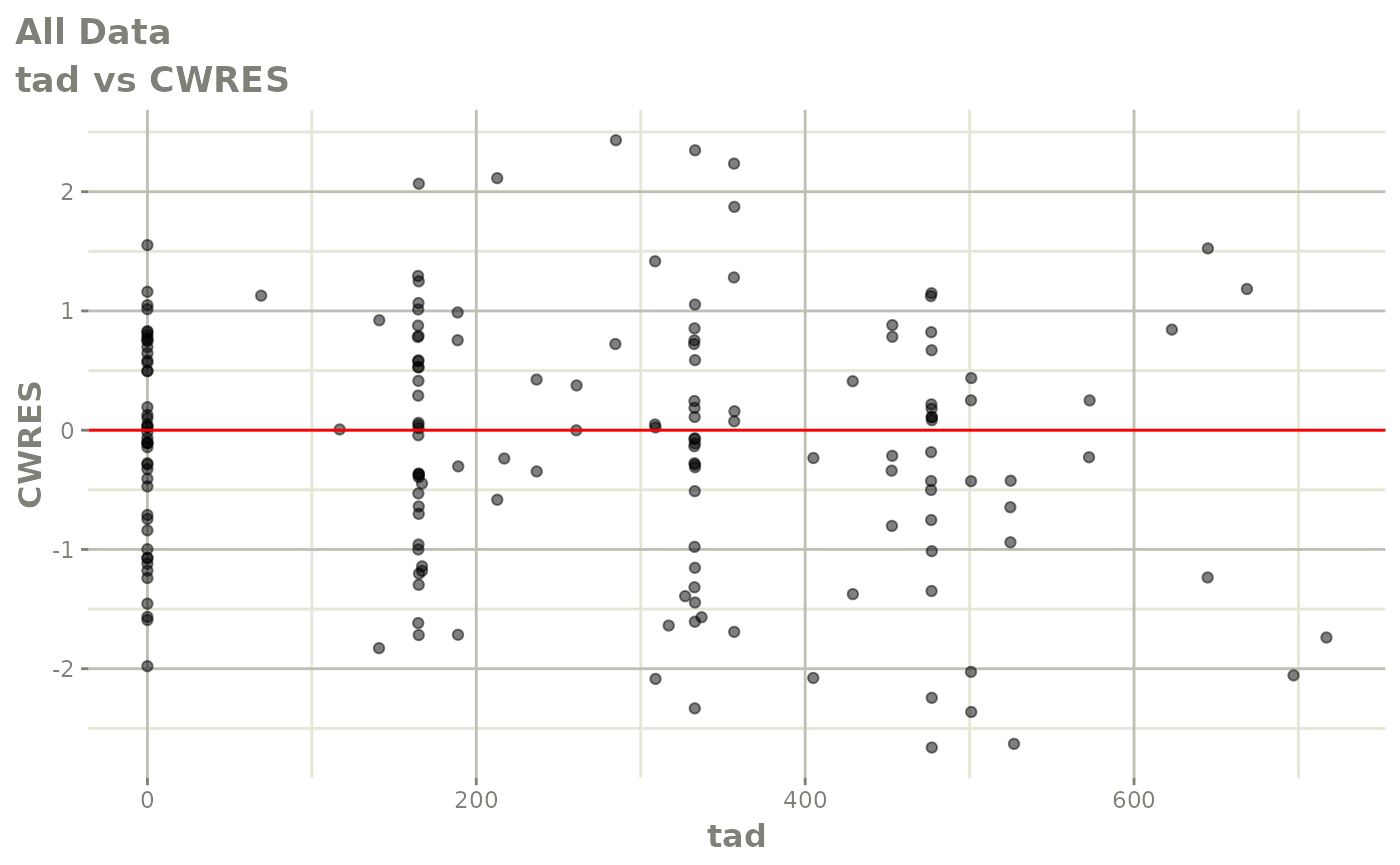
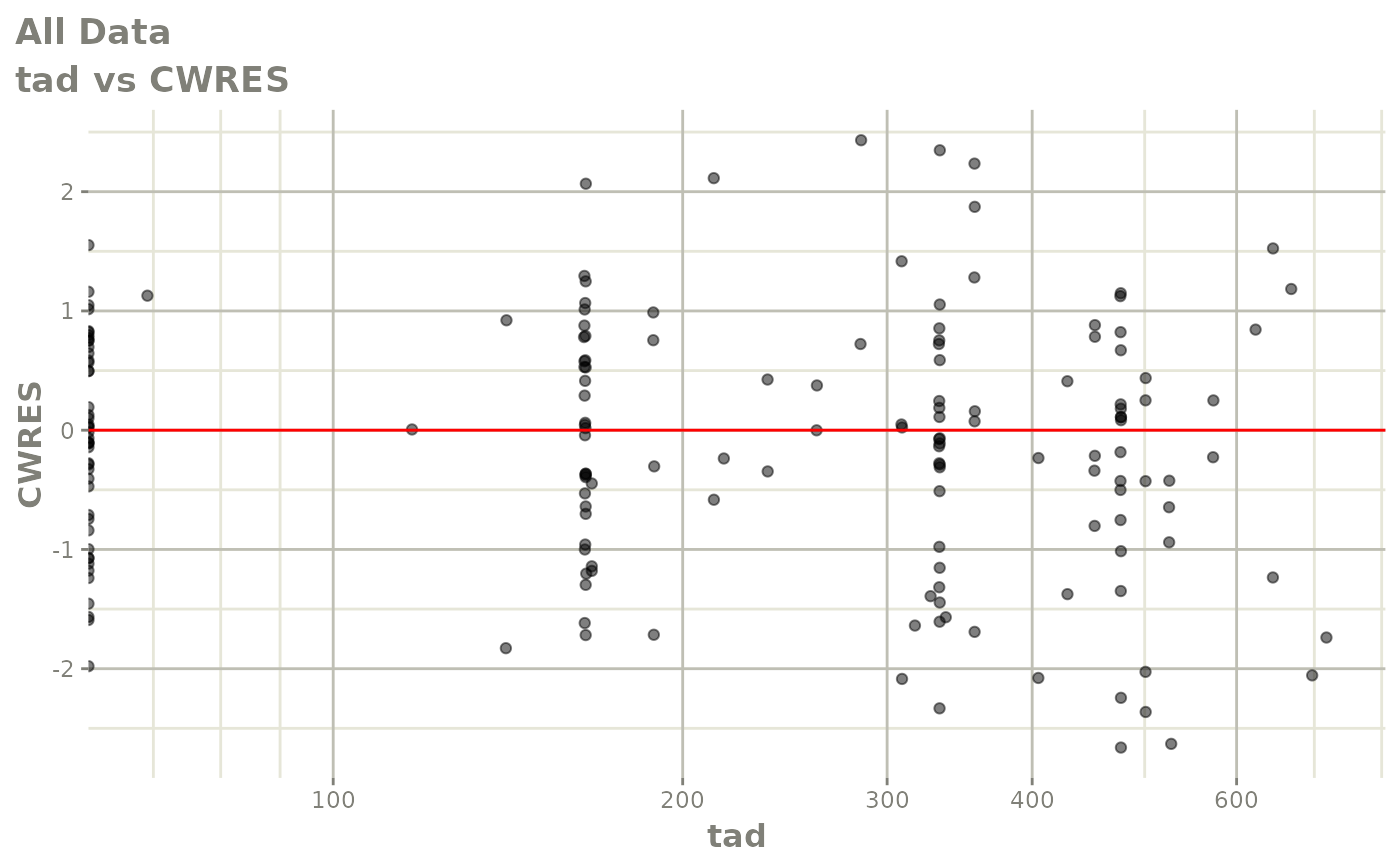
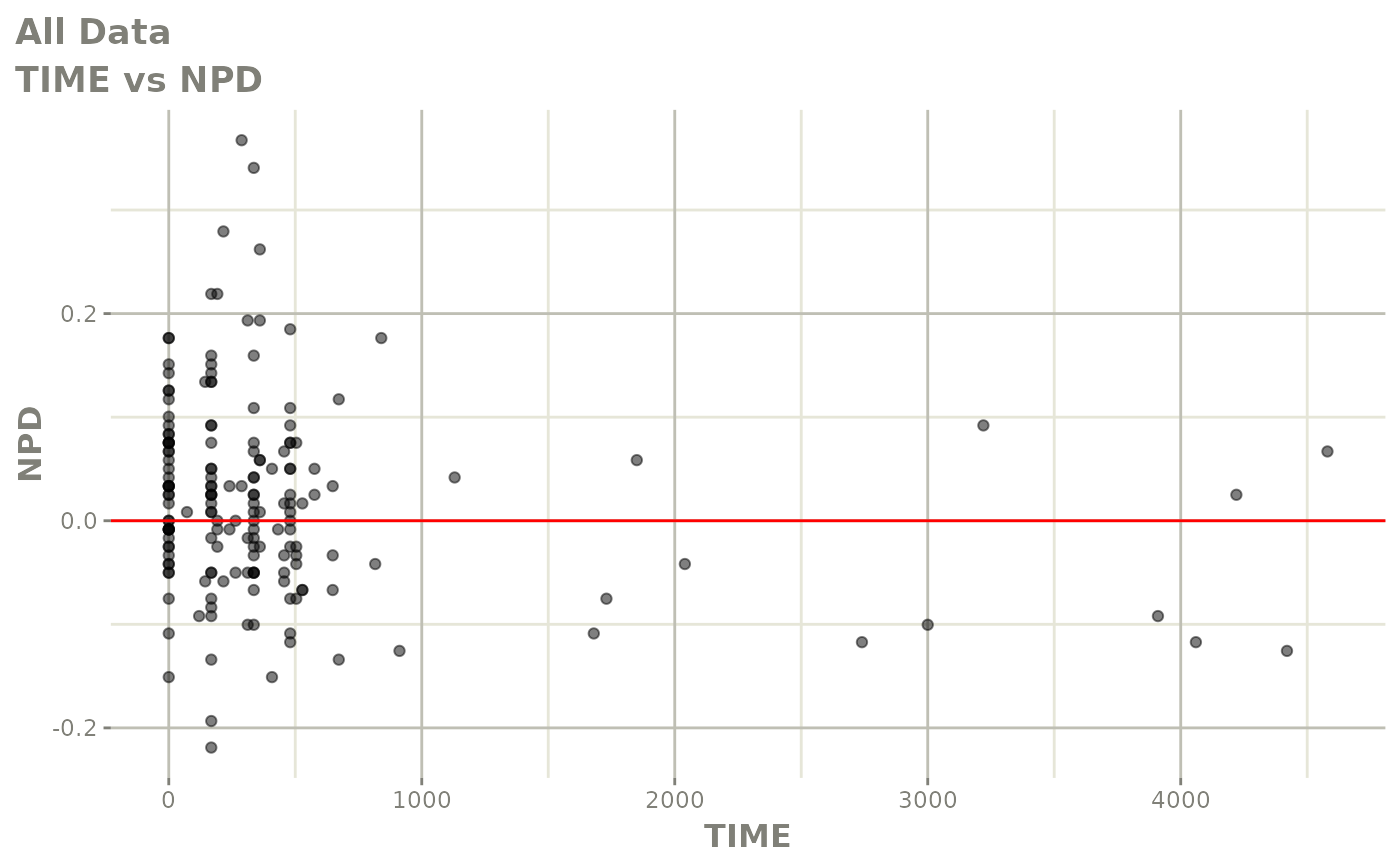
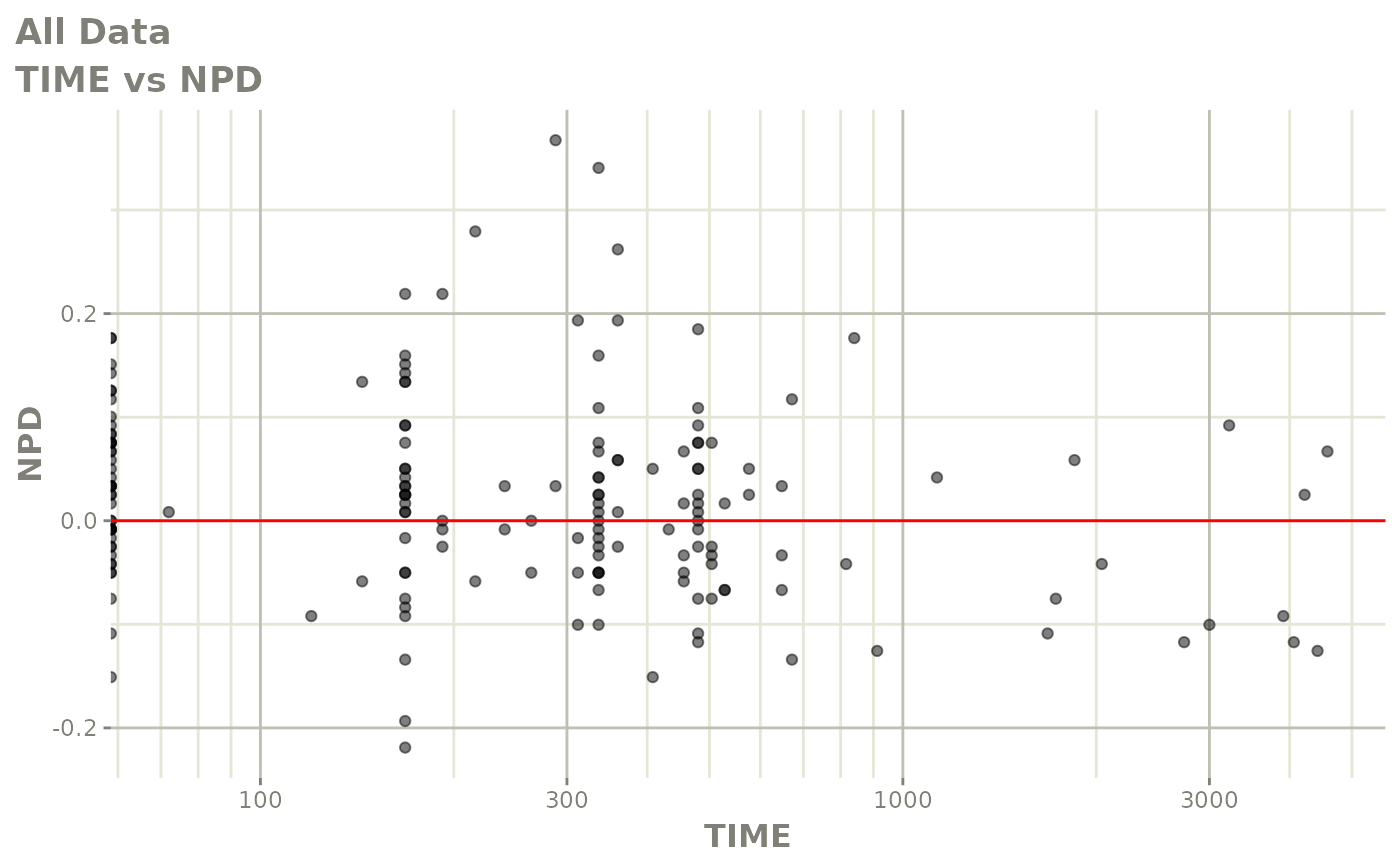
print(res_vs_idv(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Time (h)"))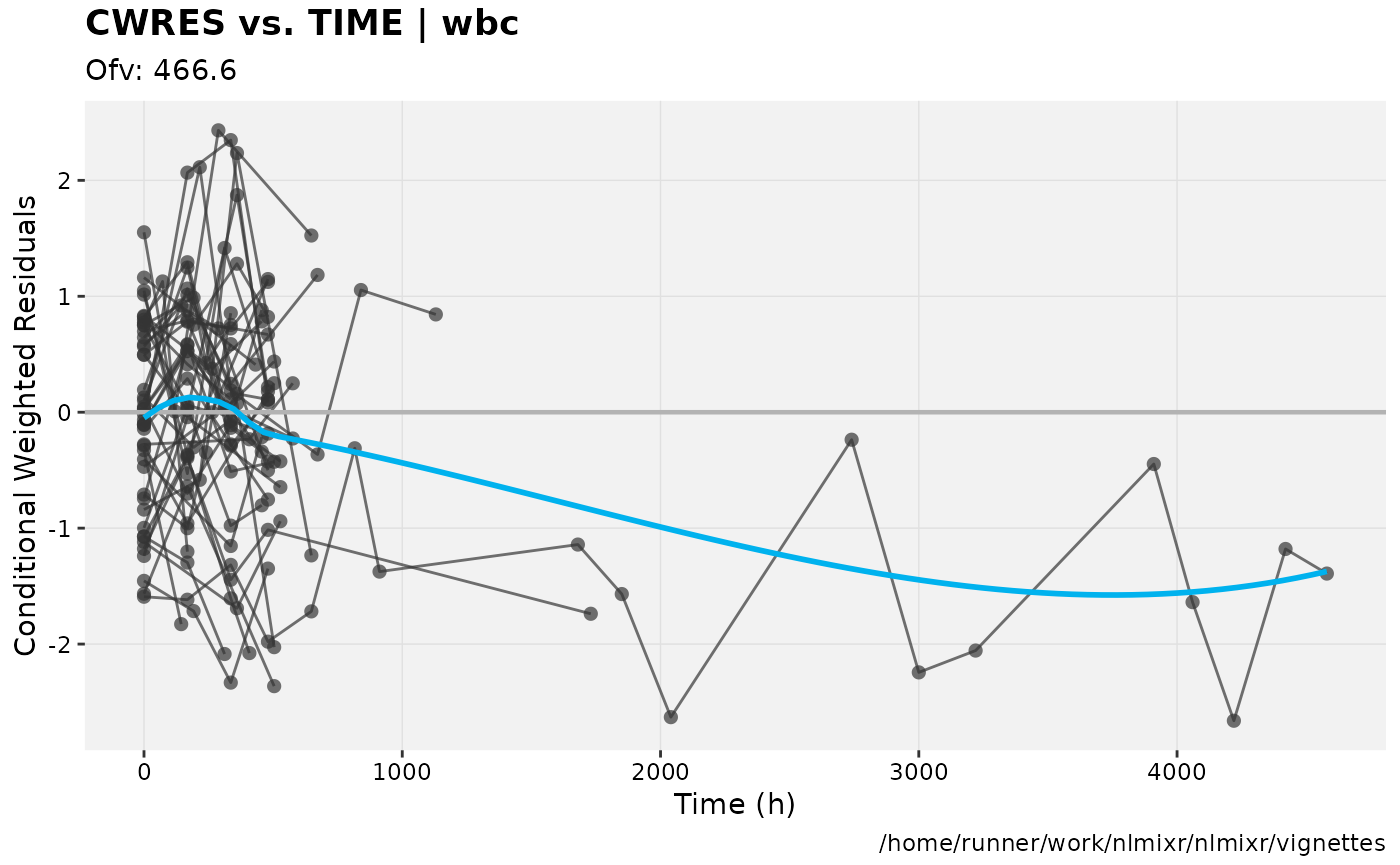
print(prm_vs_iteration(xpdb))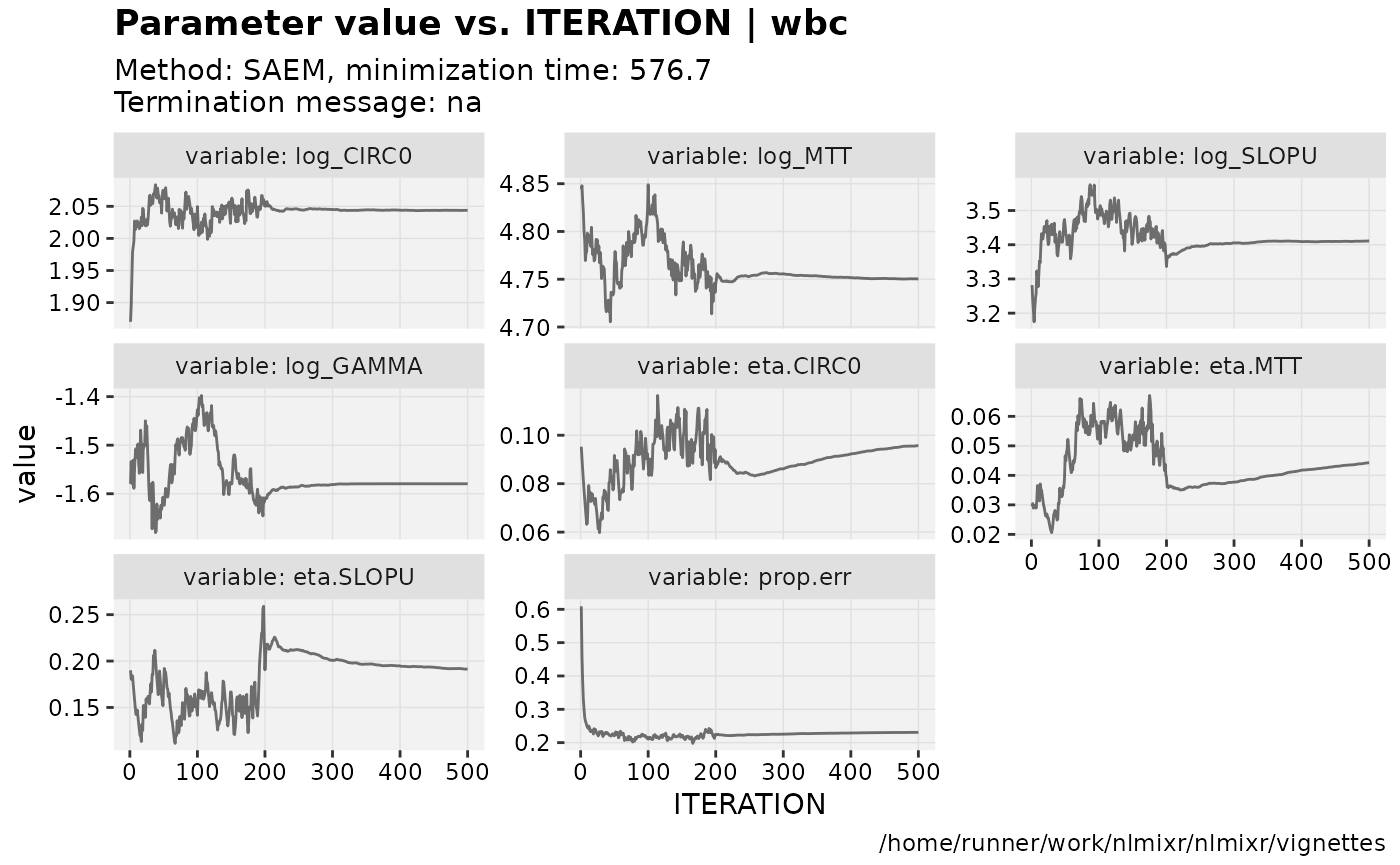
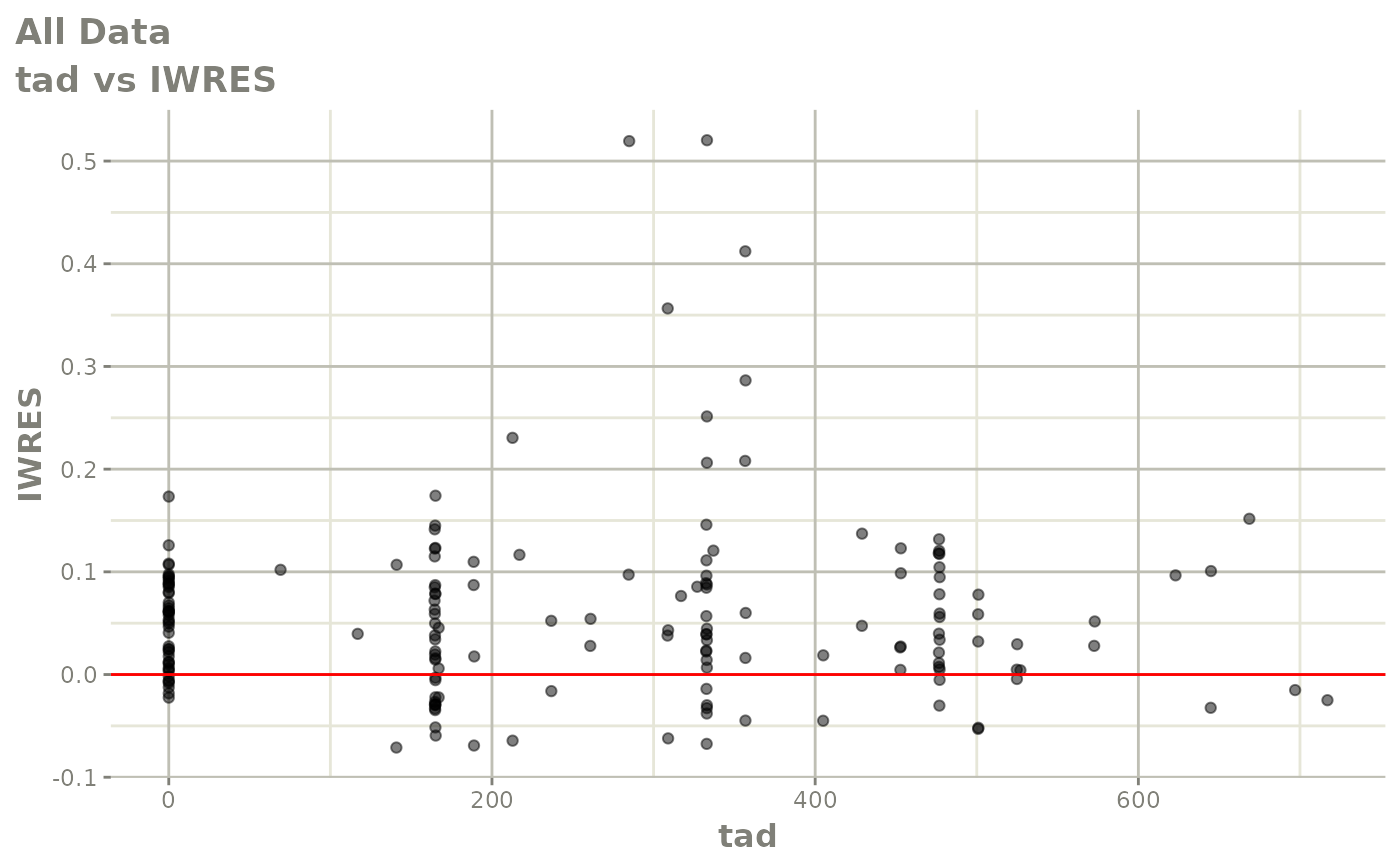
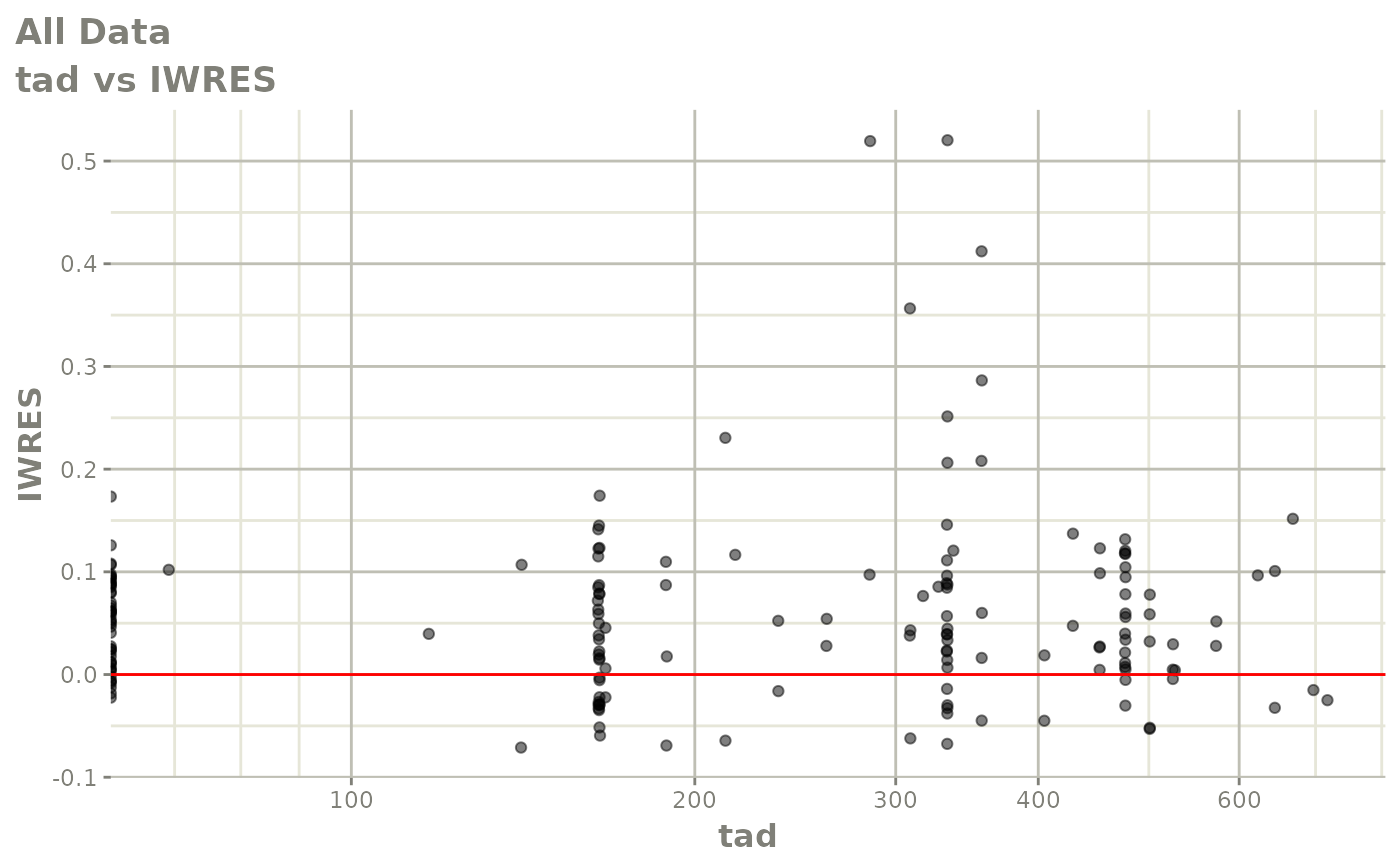
print(absval_res_vs_idv(xpdb, res = 'IWRES') +
ylab("Individual Weighted Residuals") +
xlab("Time (h)"))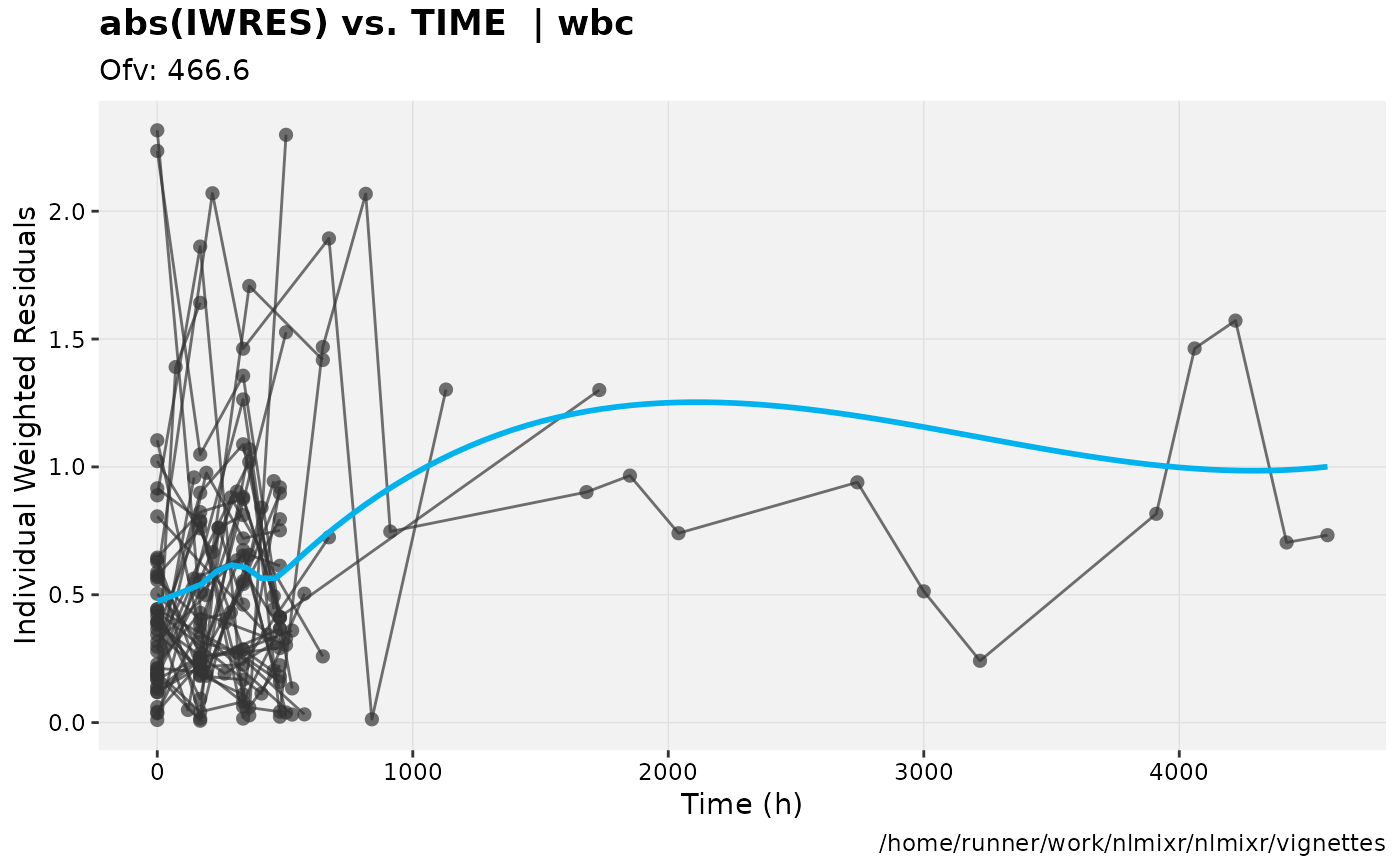
print(absval_res_vs_pred(xpdb, res = 'IWRES') +
ylab("Individual Weighted Residuals") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))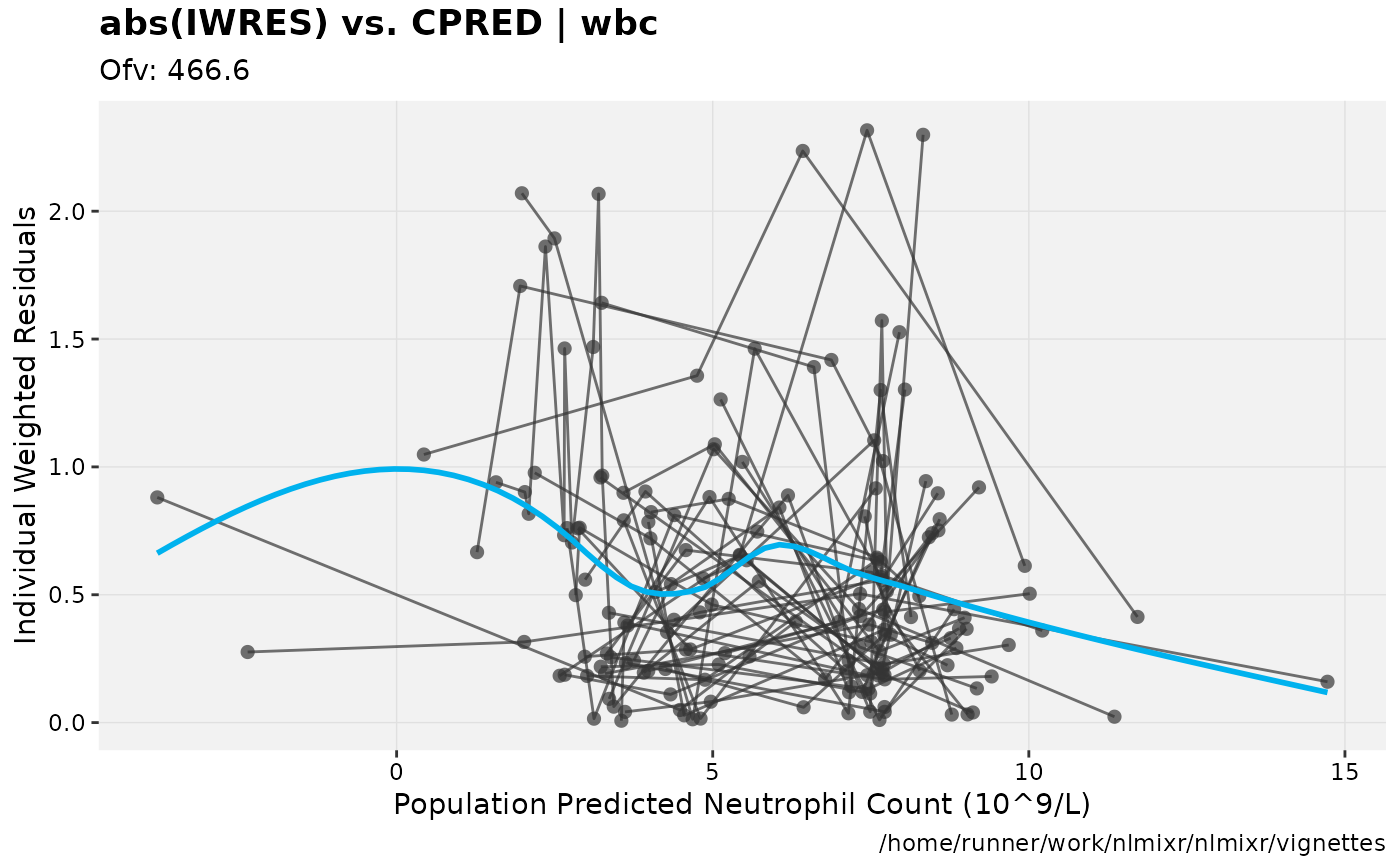
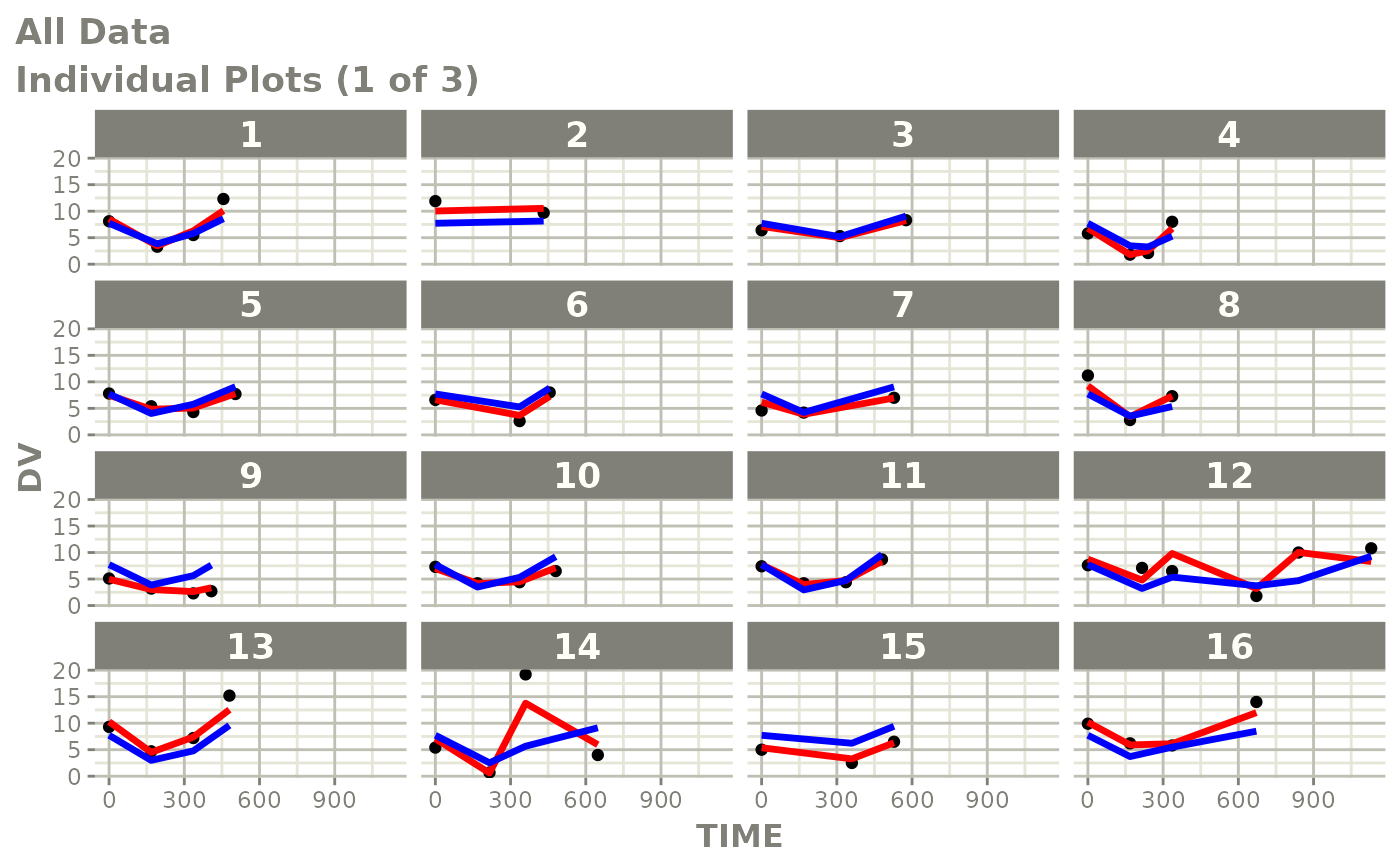
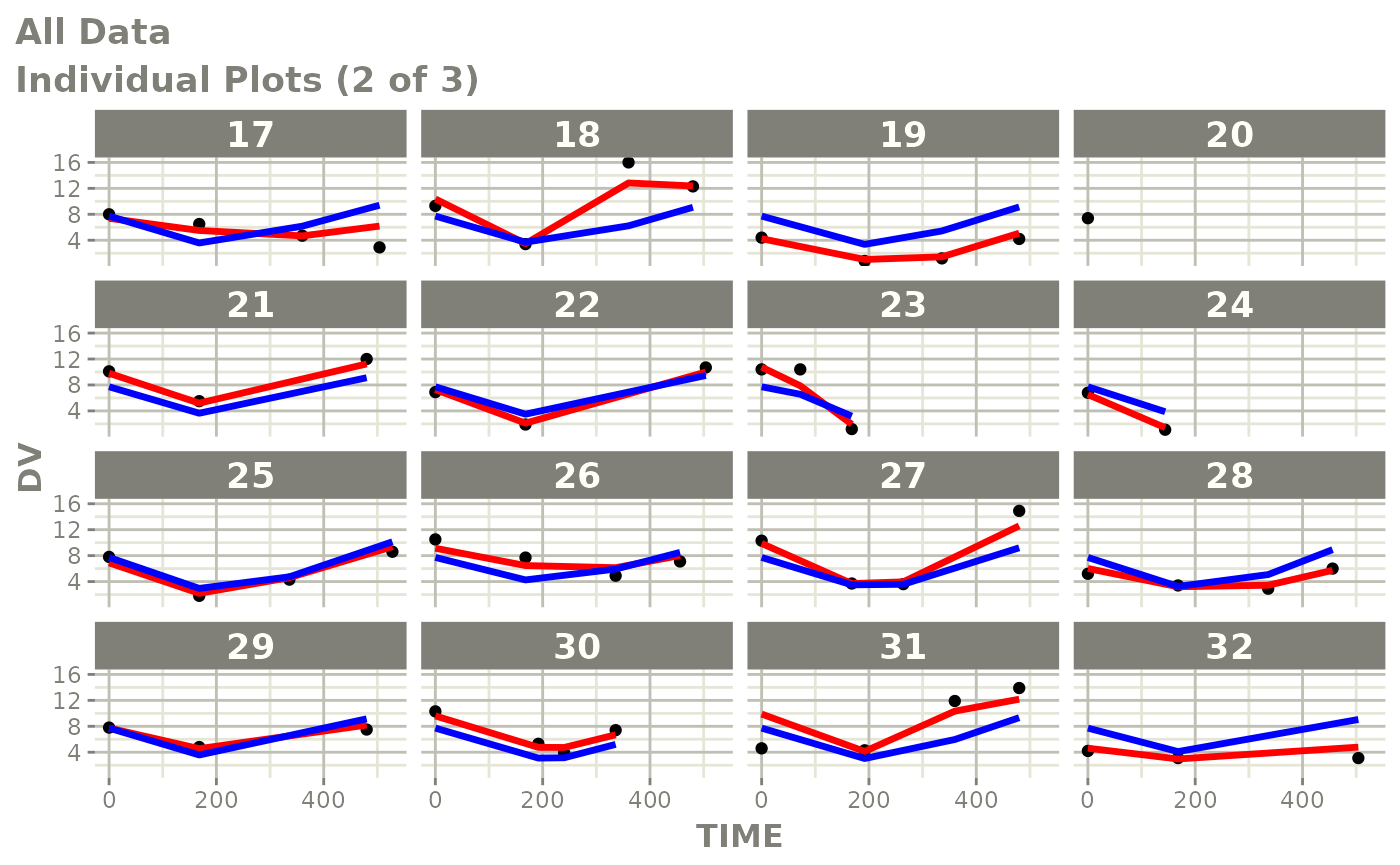
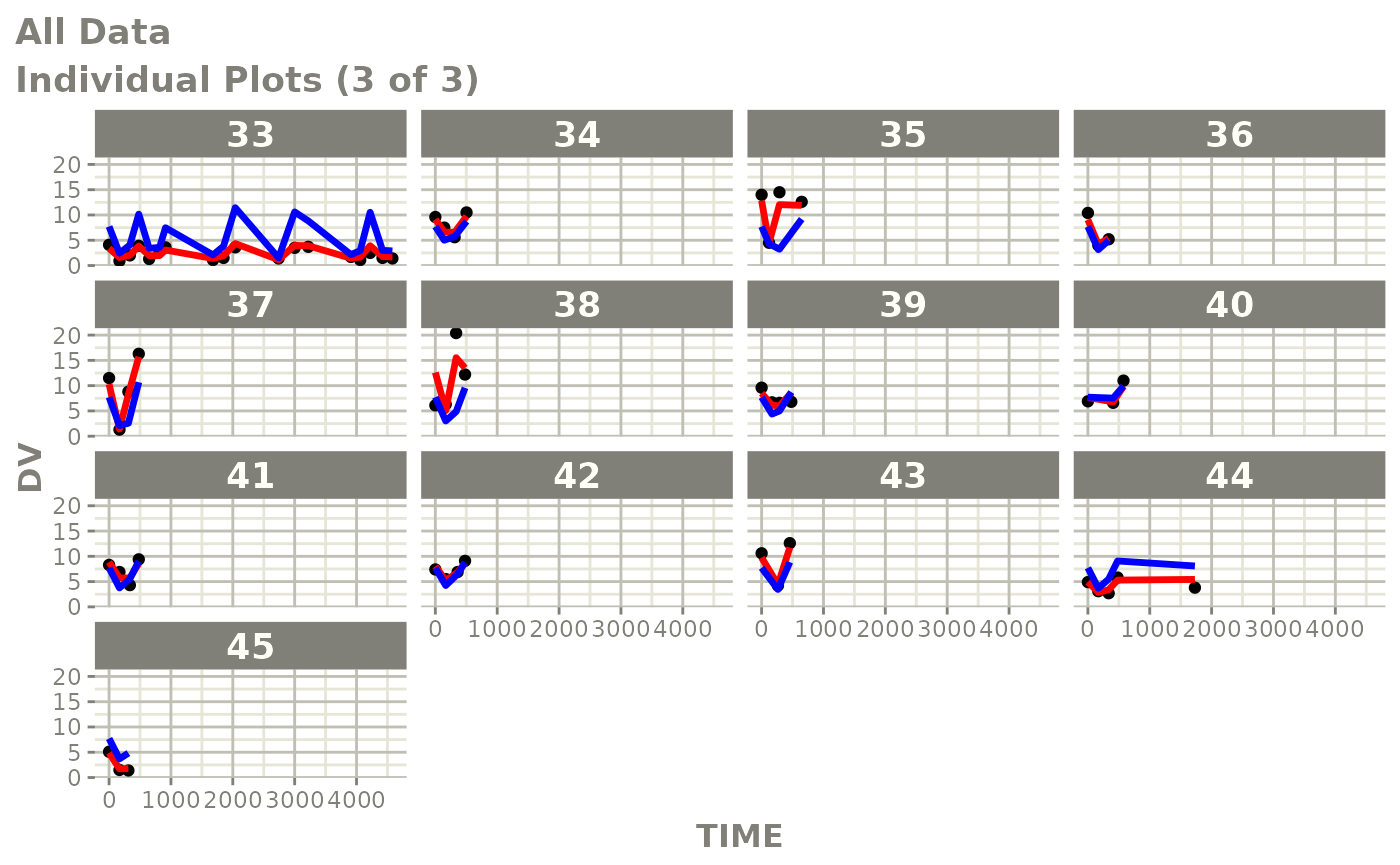
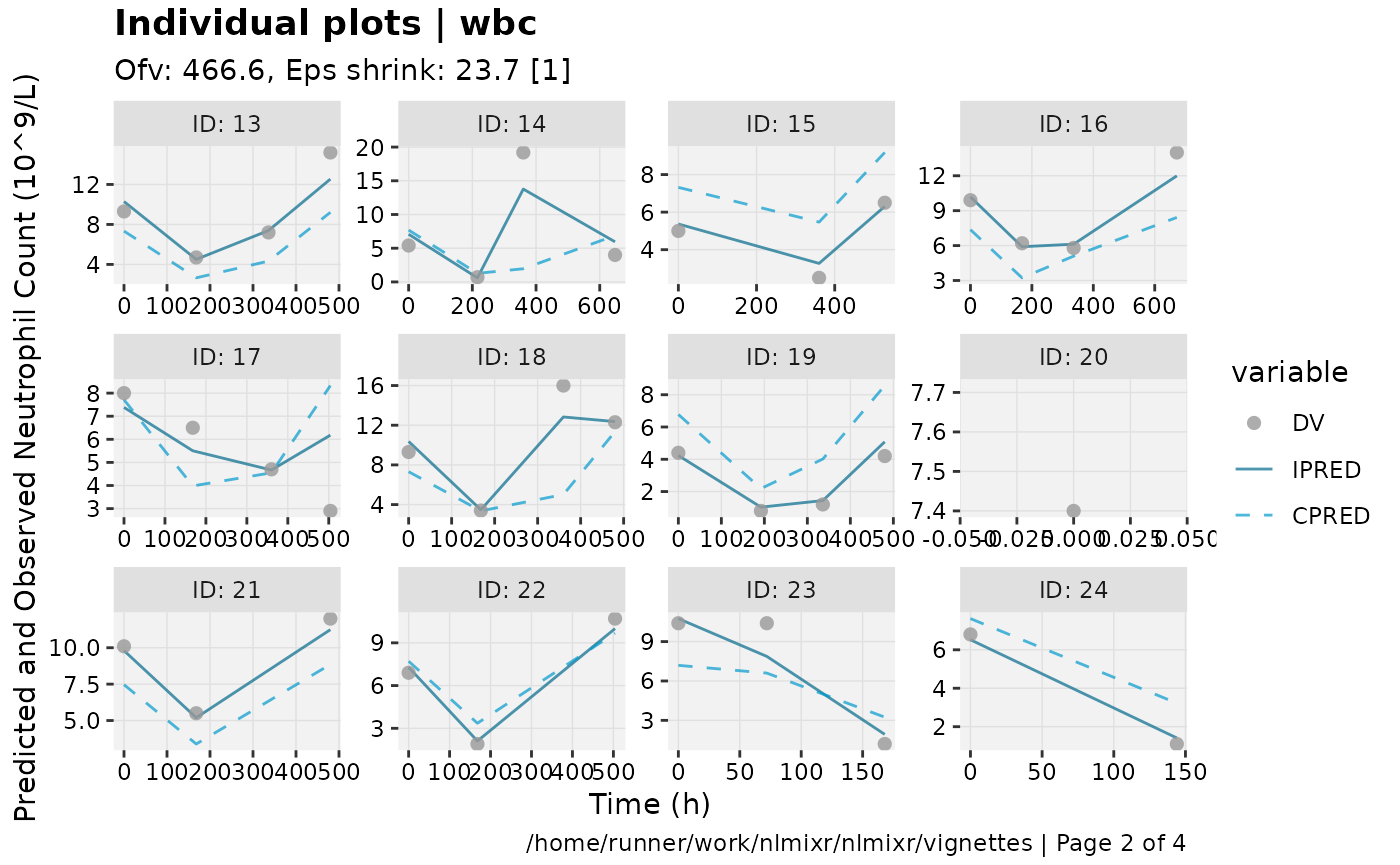
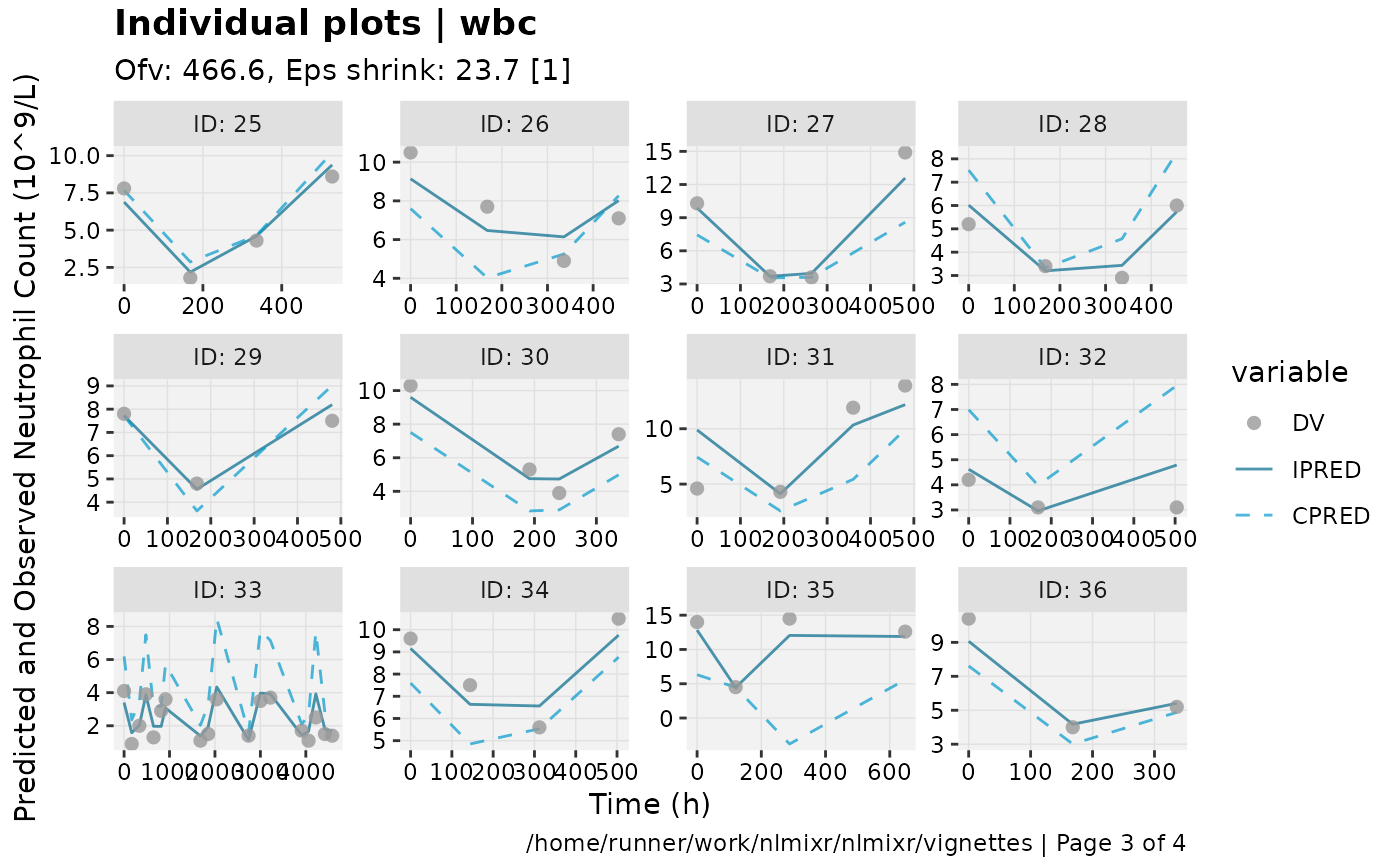
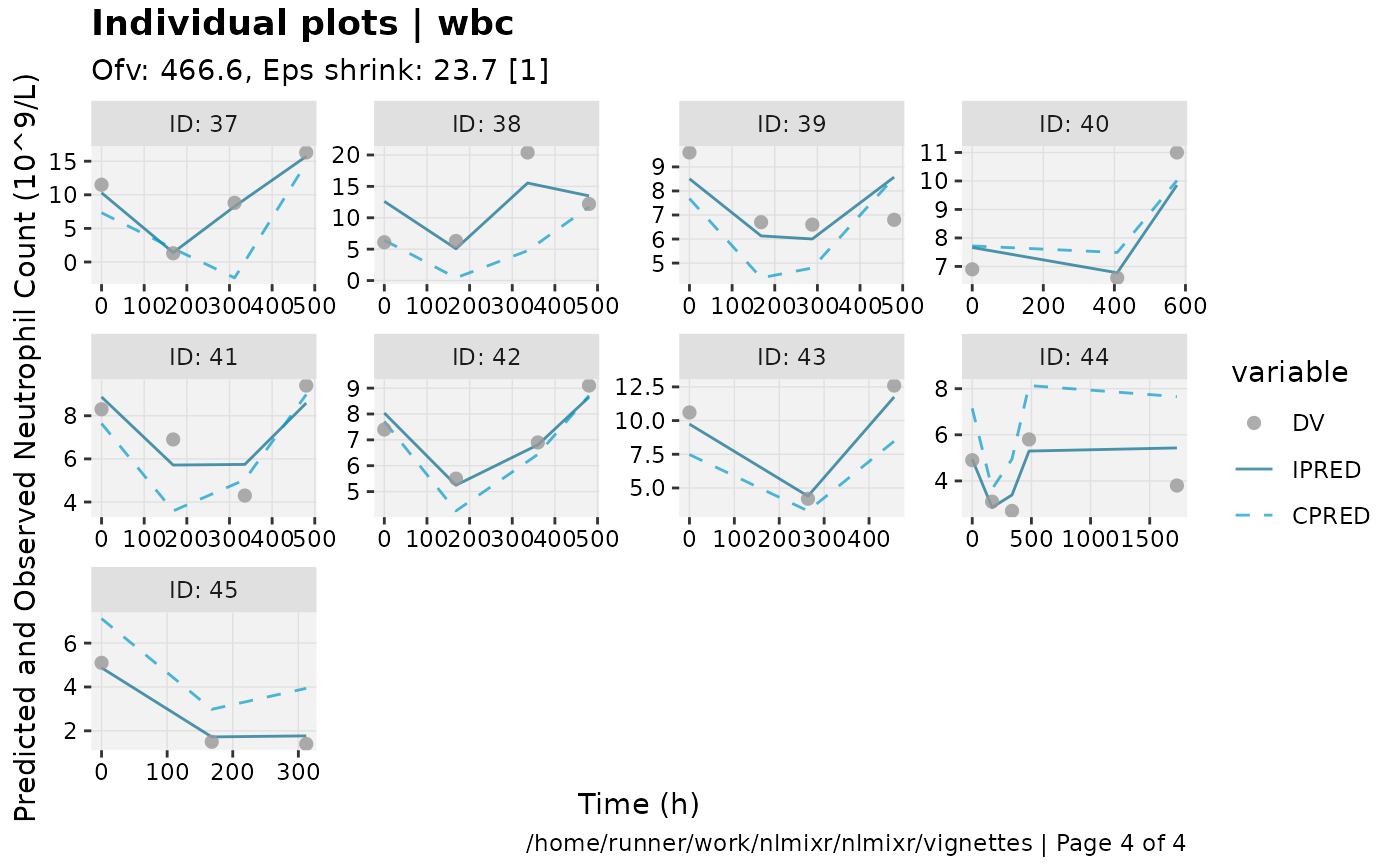
print(ind_plots(xpdb, nrow=3, ncol=4) +
ylab("Predicted and Observed Neutrophil Count (10^9/L)") +
xlab("Time (h)"))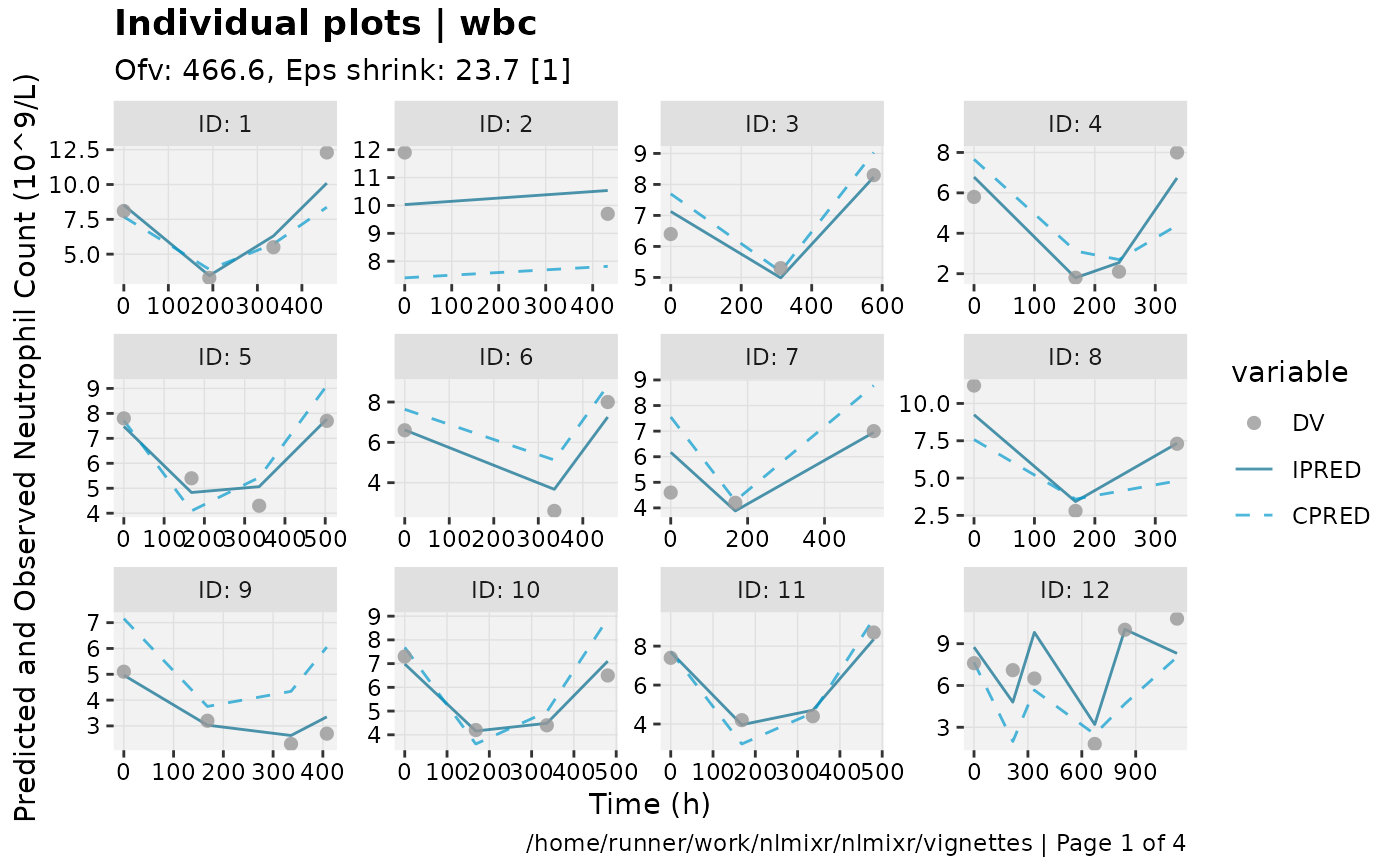
print(res_distrib(xpdb) +
ylab("Density") +
xlab("Conditional Weighted Residuals"))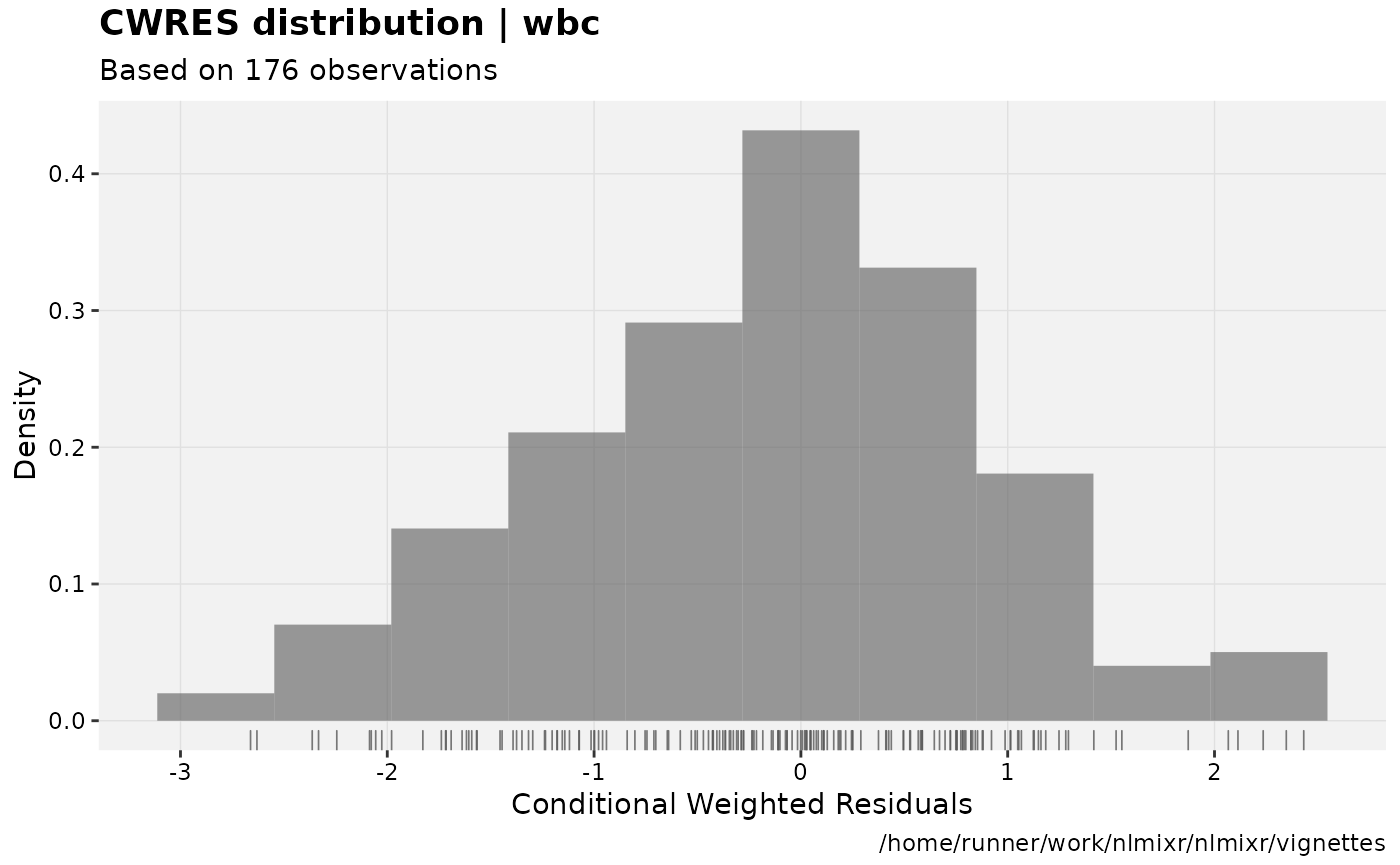
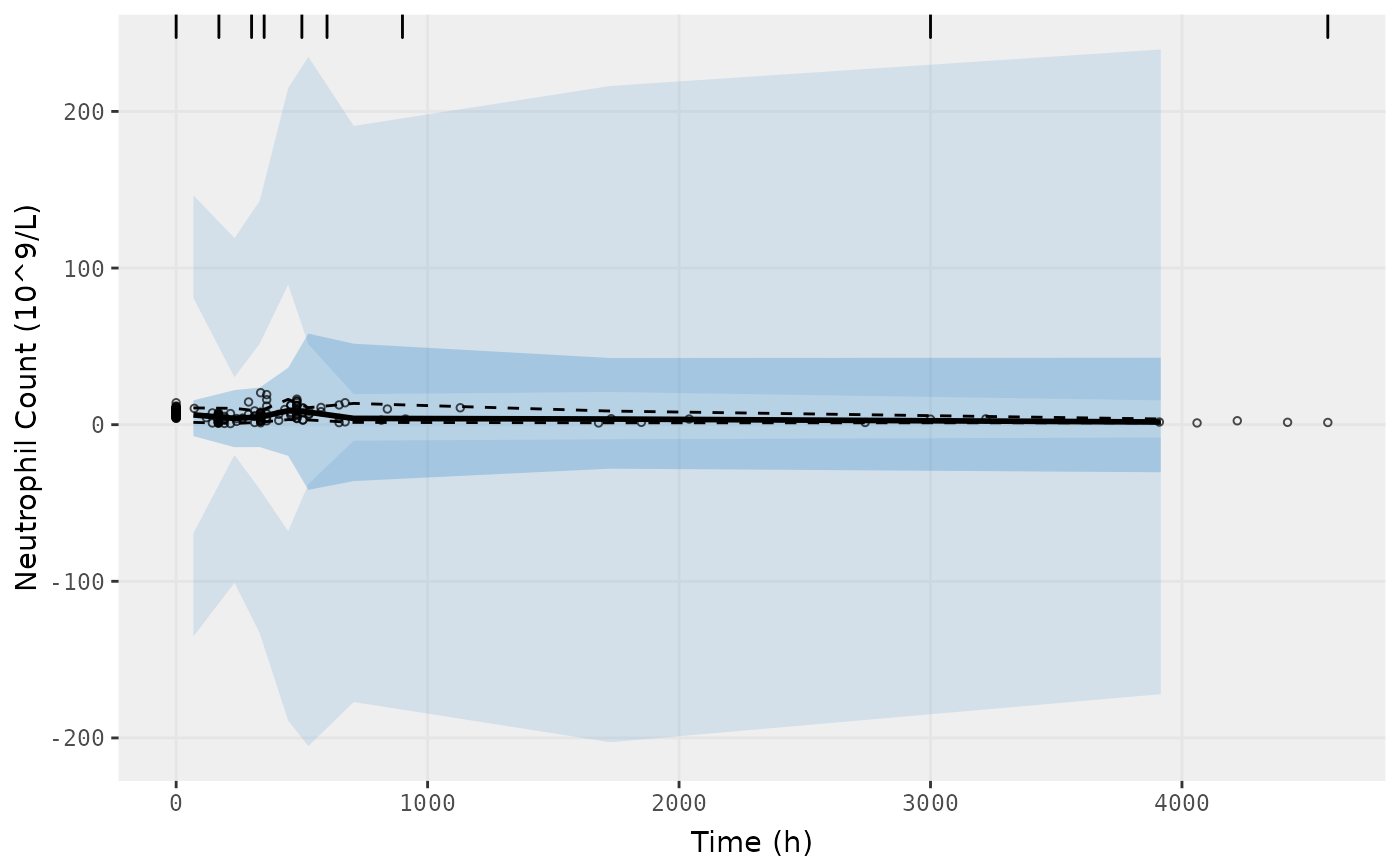
vpc.ui(fit.S, n=500, n_bins = 10, show=list(obs_dv=T), ylab = "Neutrophil Count (10^9/L)", xlab = "Time (h)")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:07
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now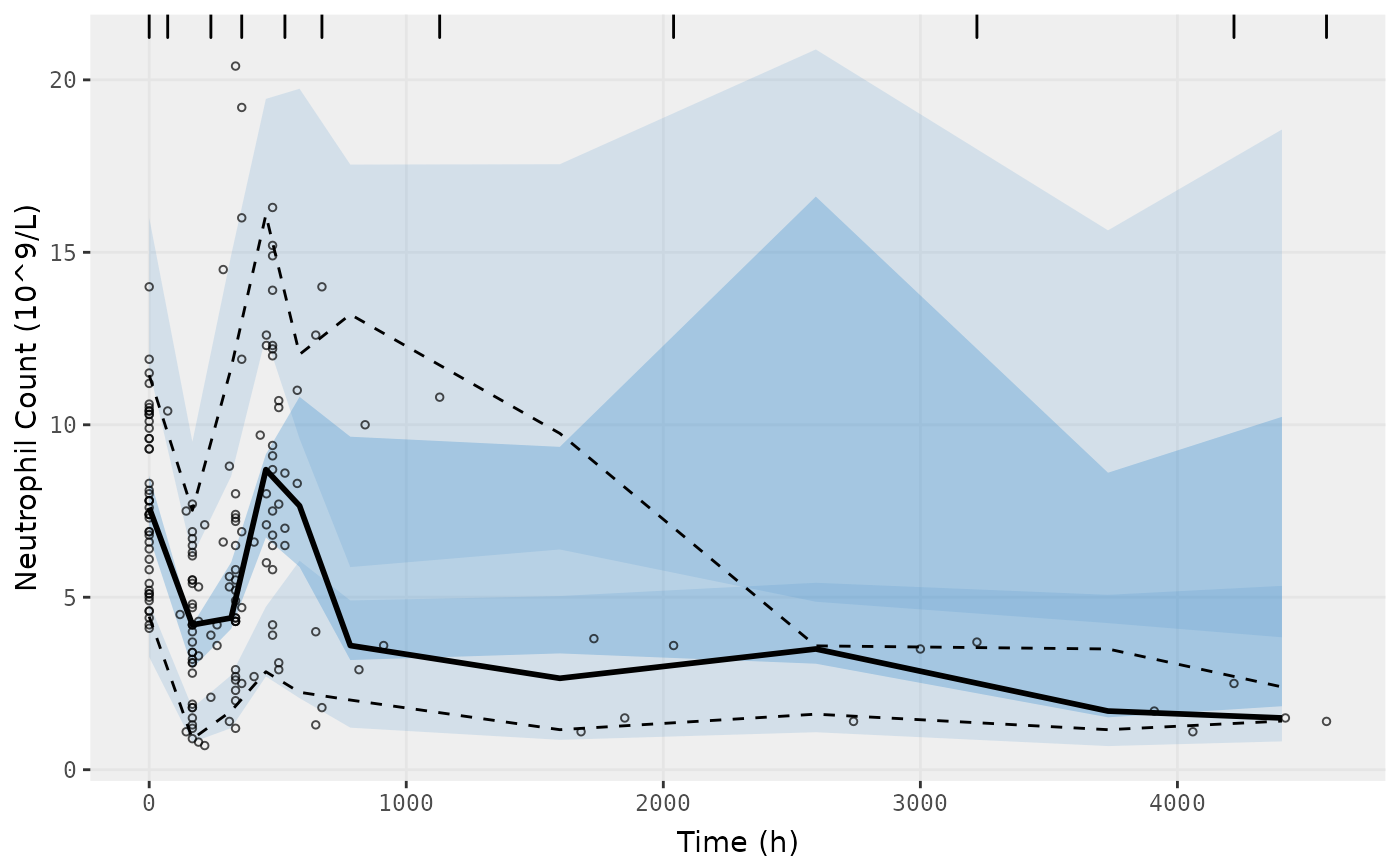
vpc.ui(fit.S, n=500, bins = c(0,170,300,350,500,600,900,3000,4580), show=list(obs_dv=T), ylab = "Neutrophil Count (10^9/L)", xlab = "Time (h)")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:07
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now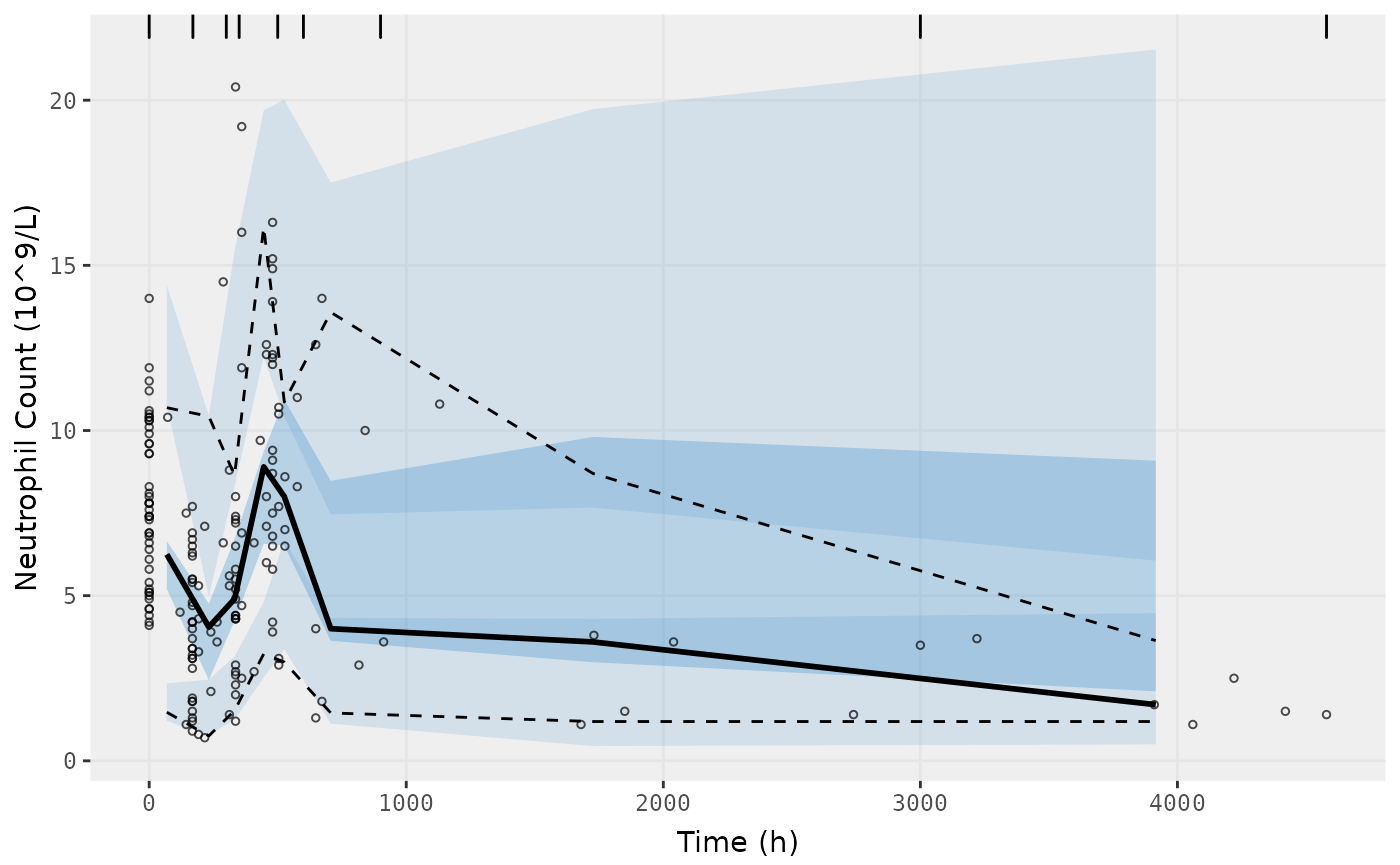
Fit model using FOCEi
fit.F <- nlmixr(wbc, d3, est="focei", list(print=0), table=list(cwres=TRUE, npde=TRUE))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [1] "V2I" "CLI" "V1I"
#> unhandled error message: EE:[lsoda] 500000 steps taken before reaching tout
#> @(lsoda.c:750
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|===
S matrix calculation failed; Switch to R-matrix covariance.
#> =|====] 0:00:34
#> done
#> [====|====|====|====|====|====|====|====|====|====] 0:00:03FOCEi Goodness of fit plots
xpdb <- xpose_data_nlmixr(fit.F)
plot(fit.F)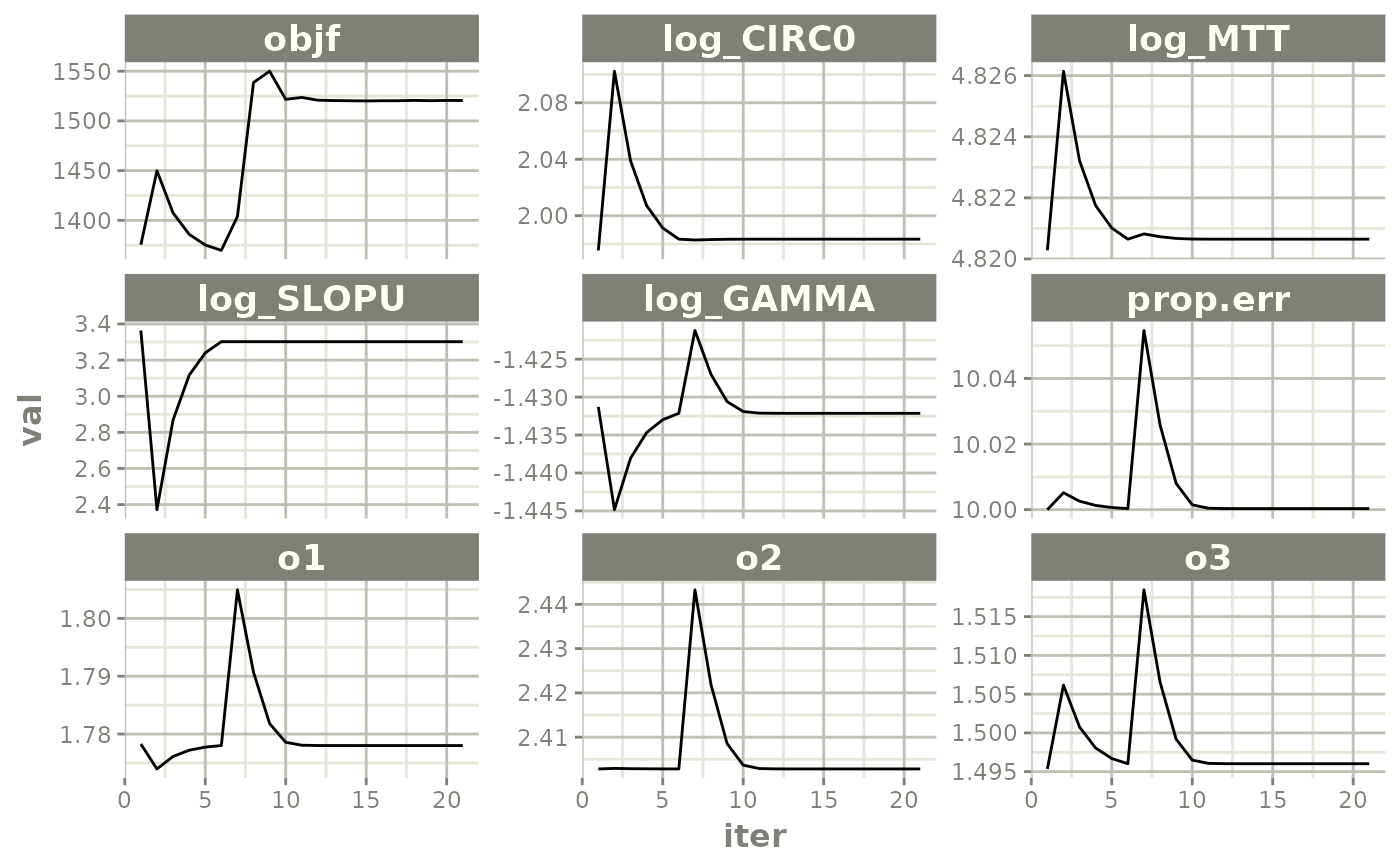
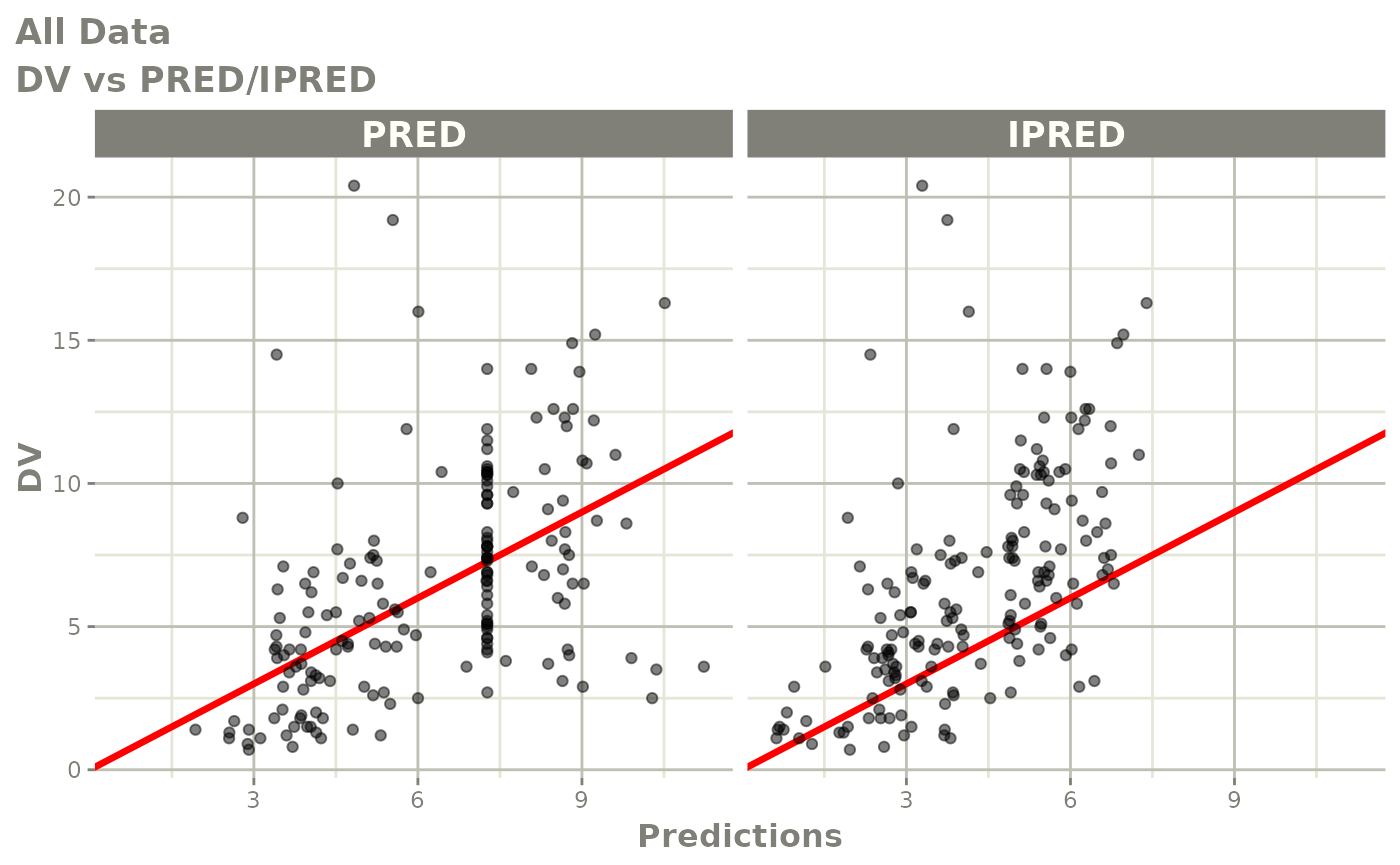
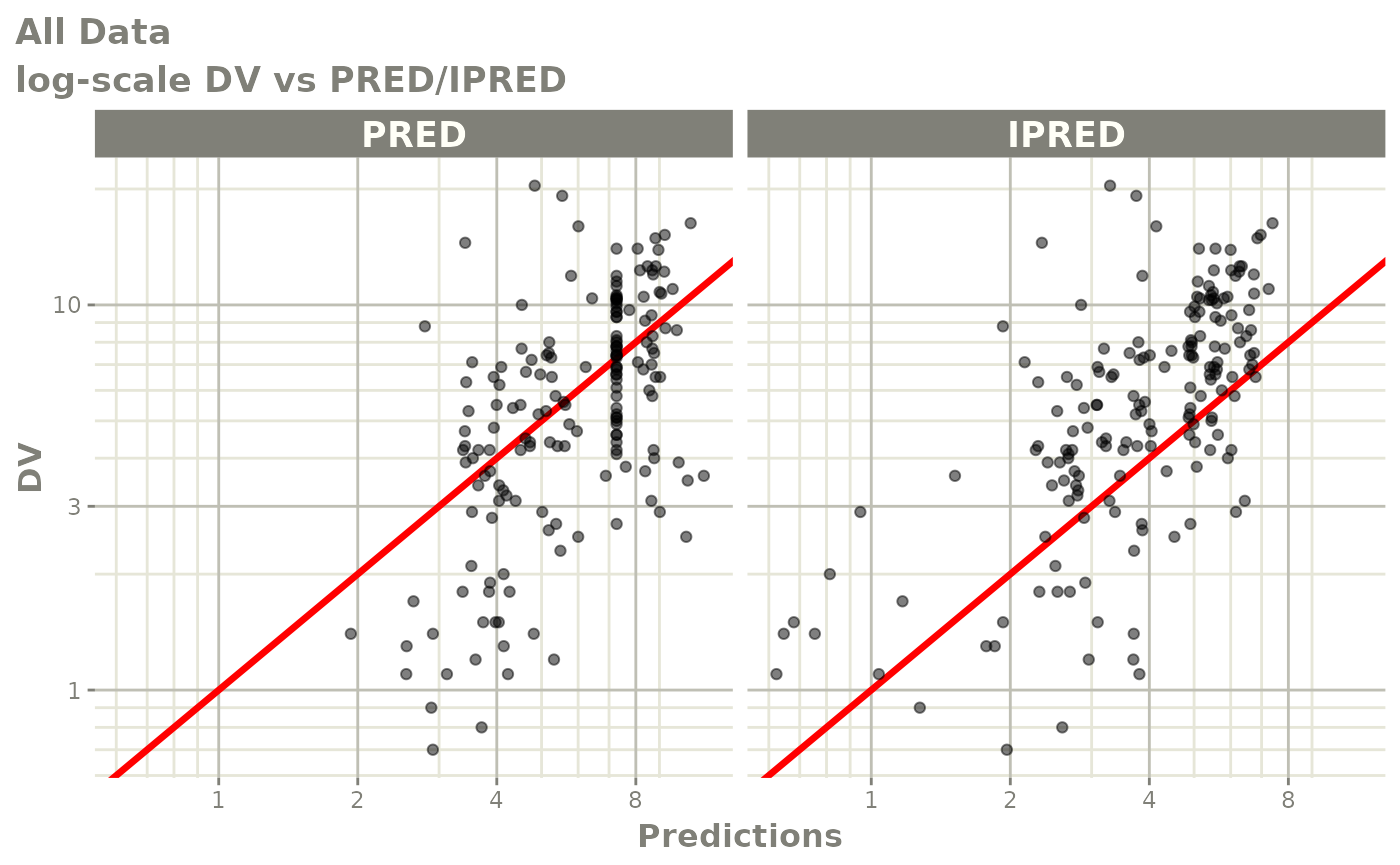
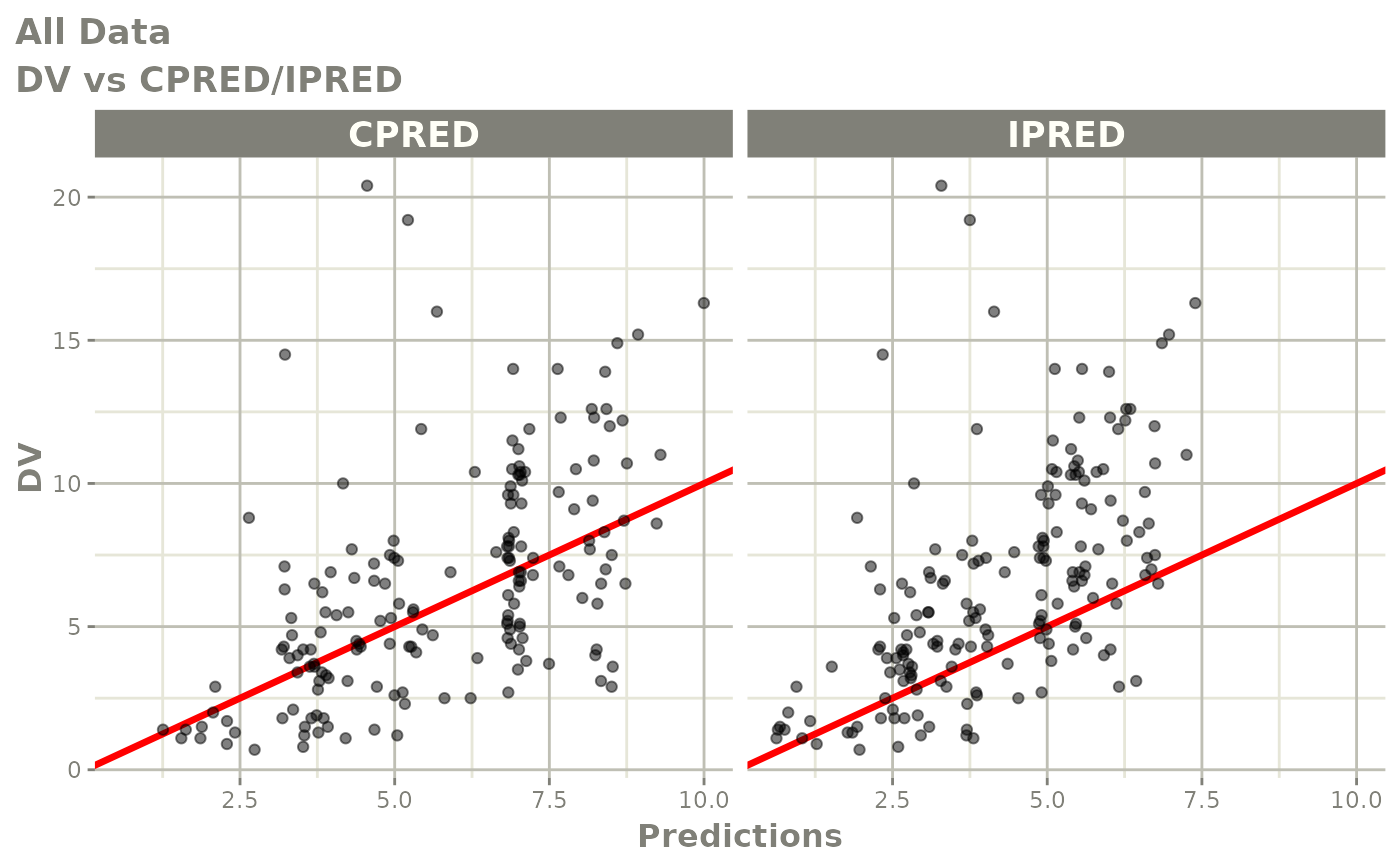
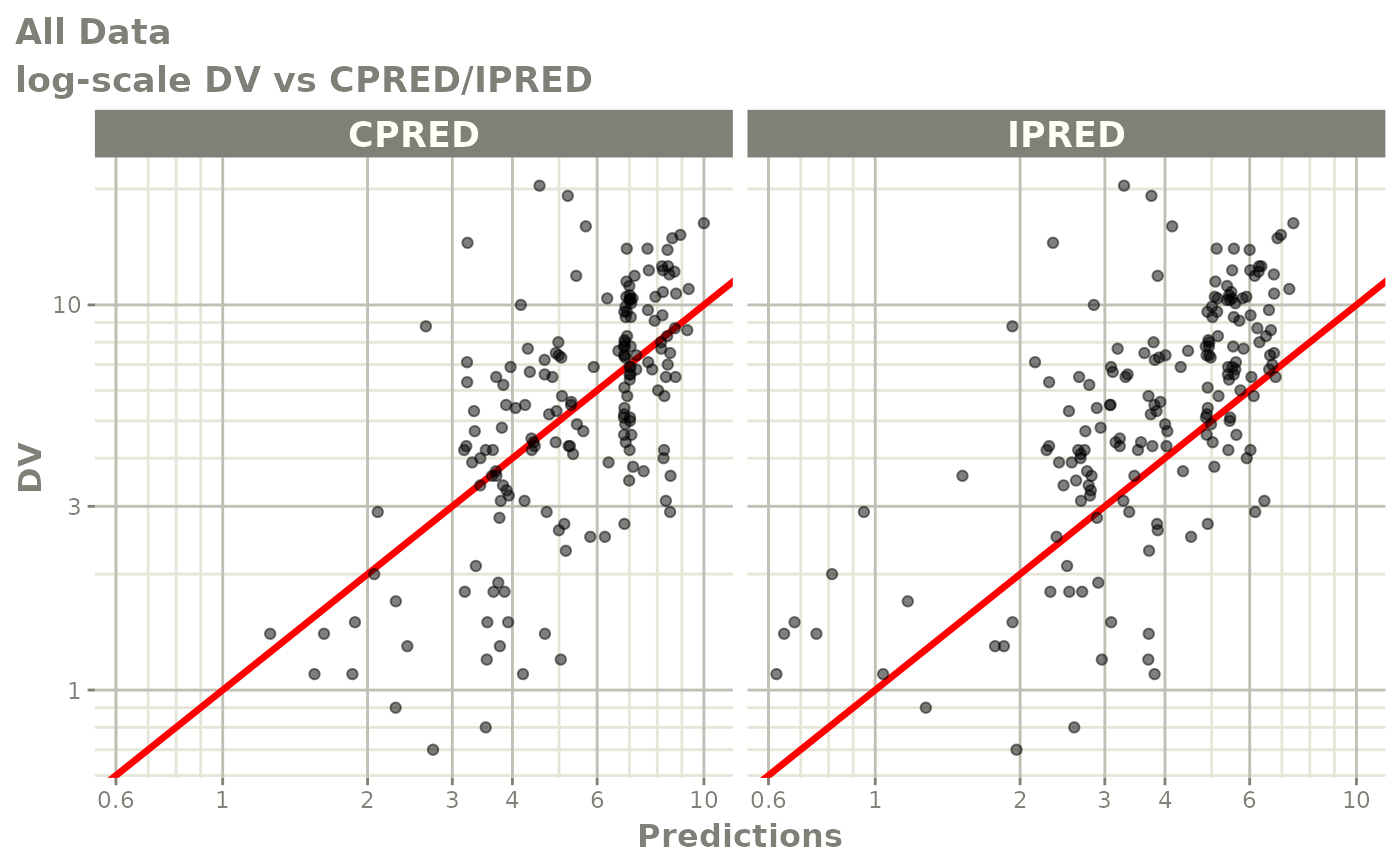
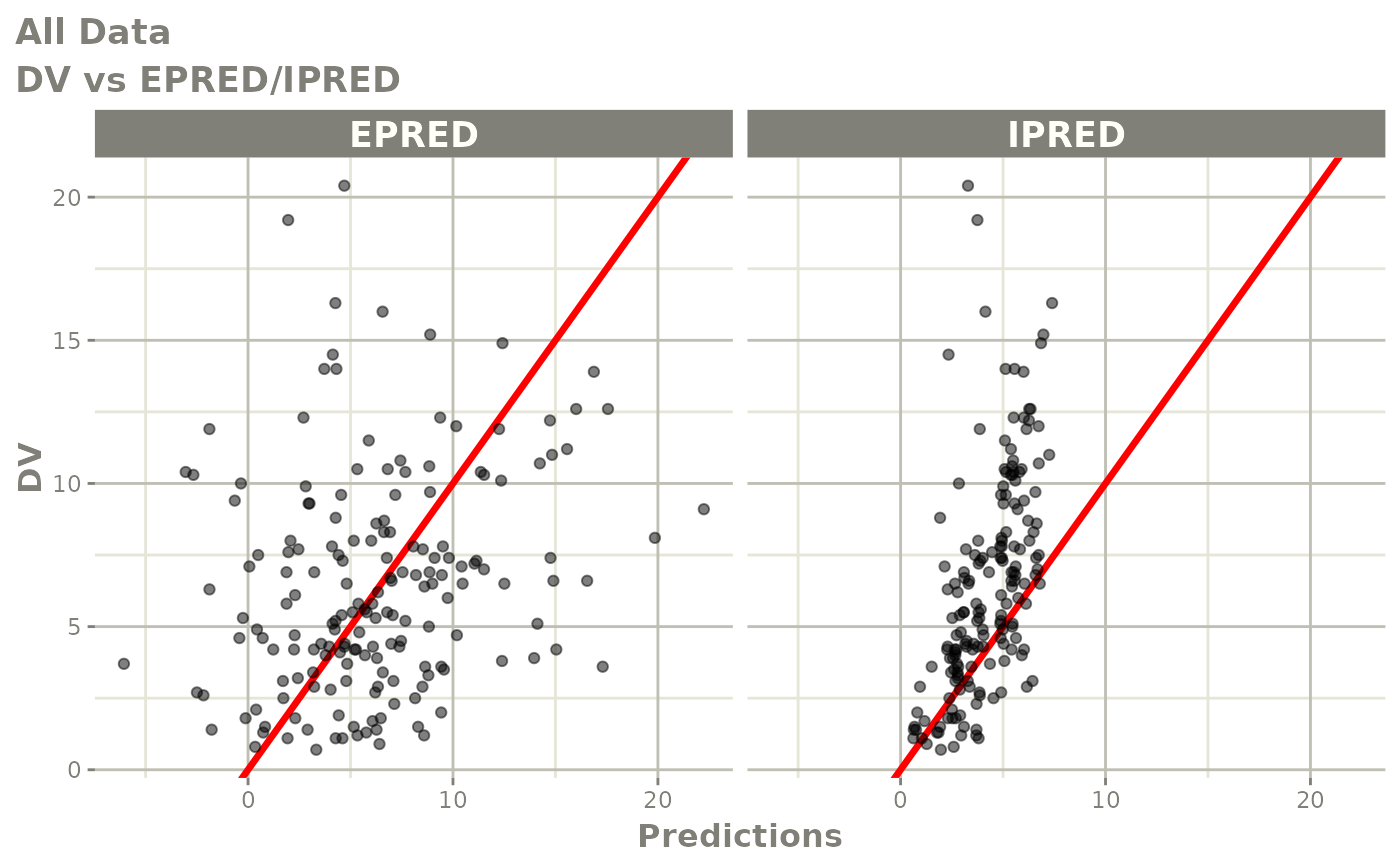
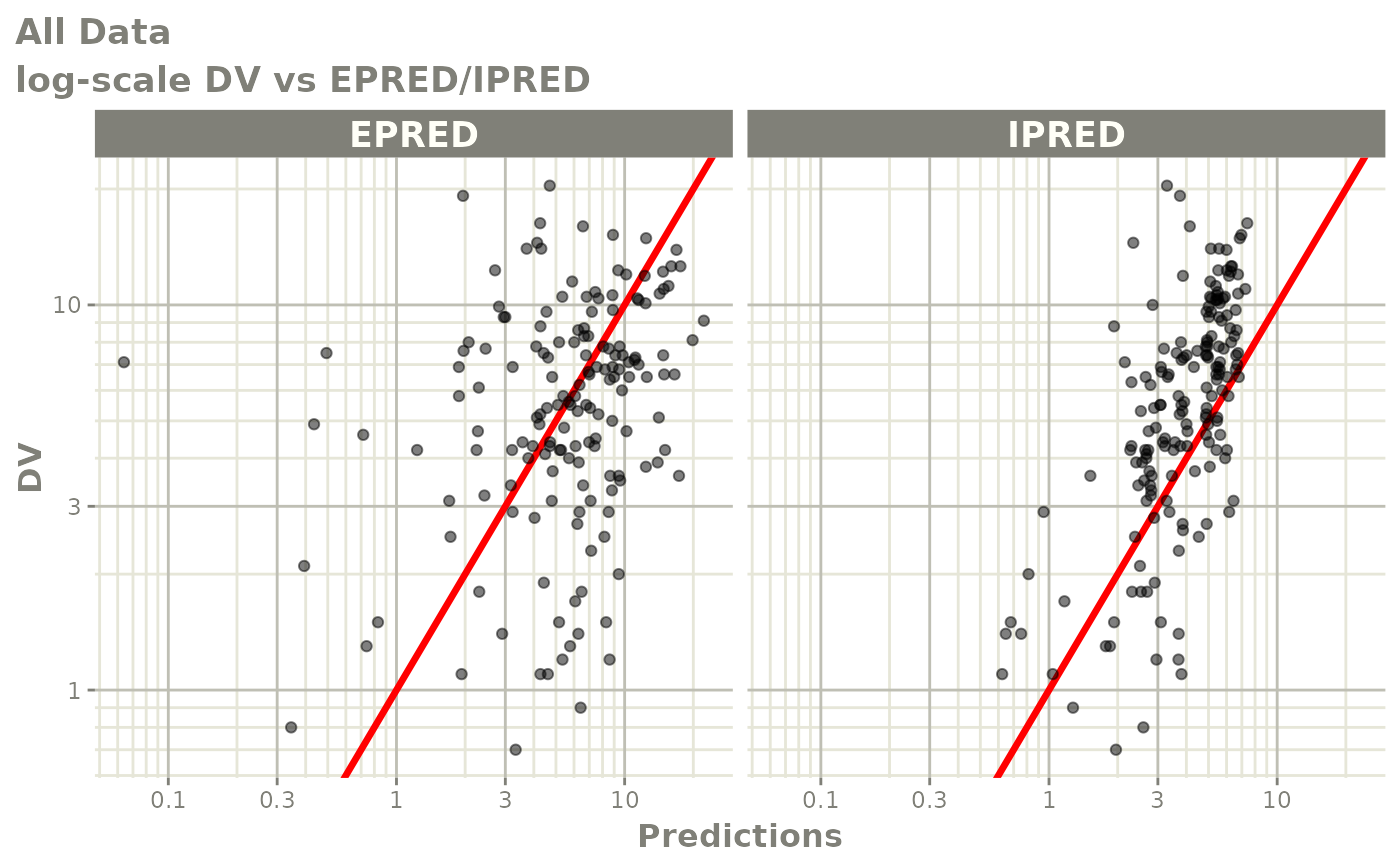
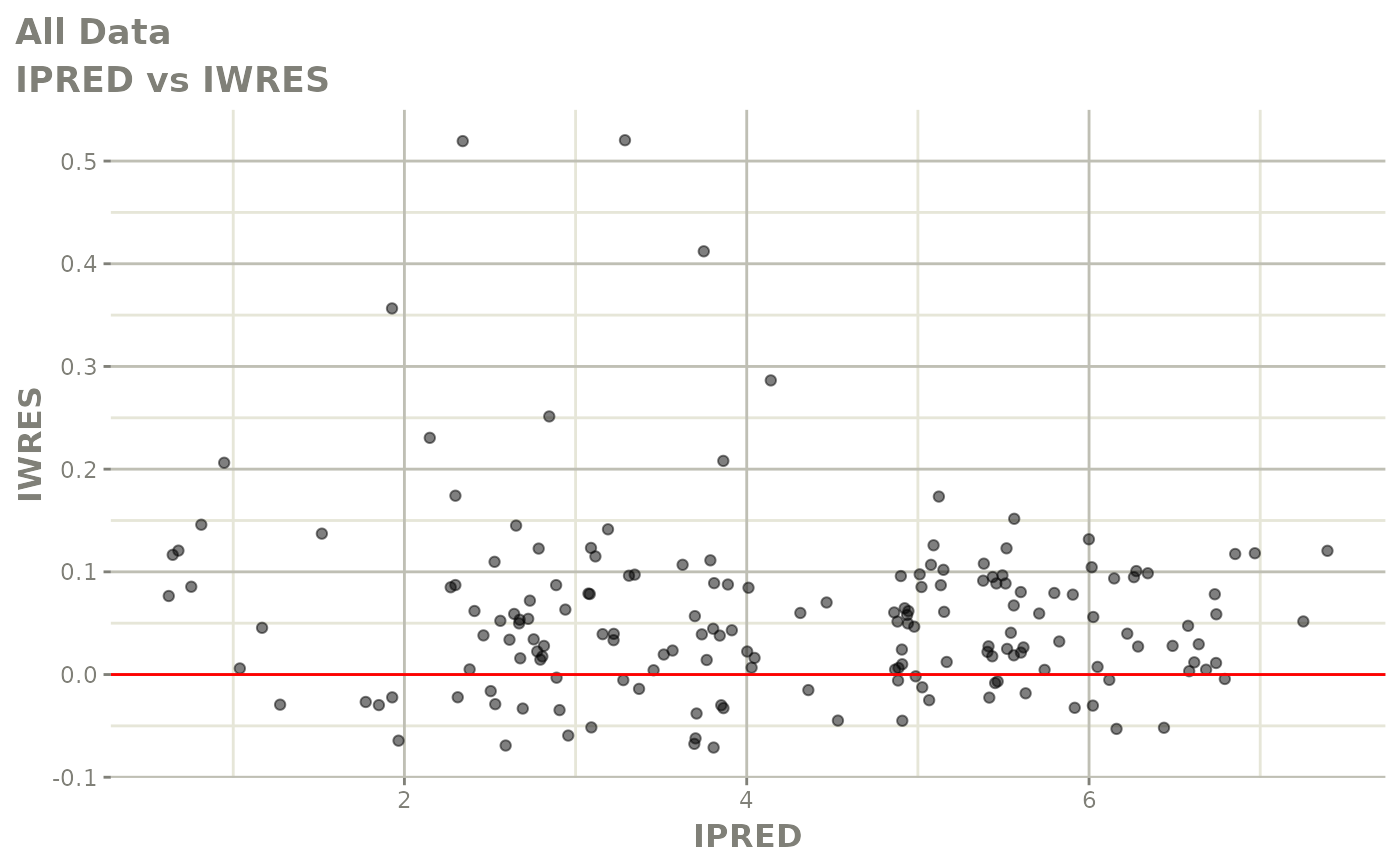
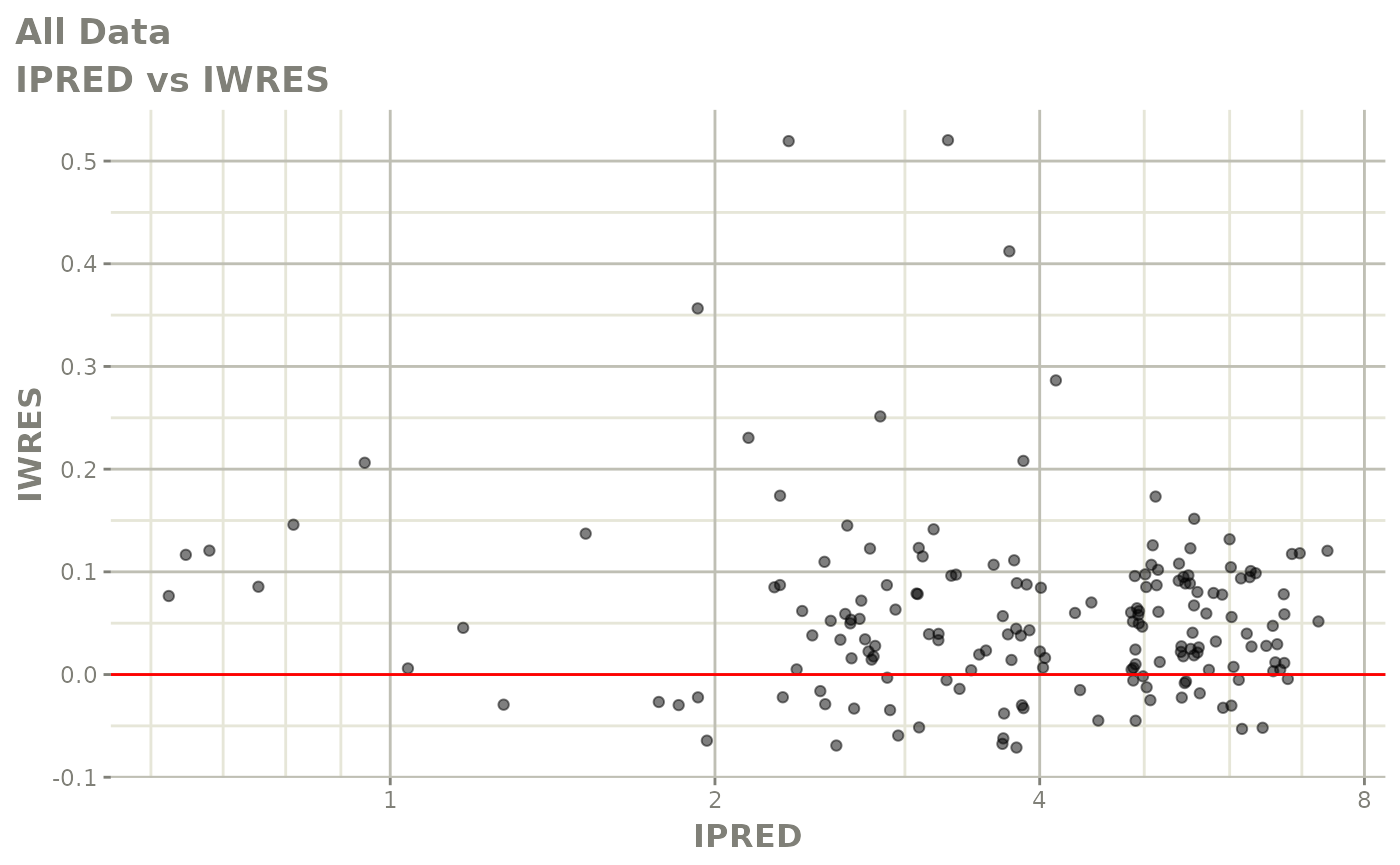
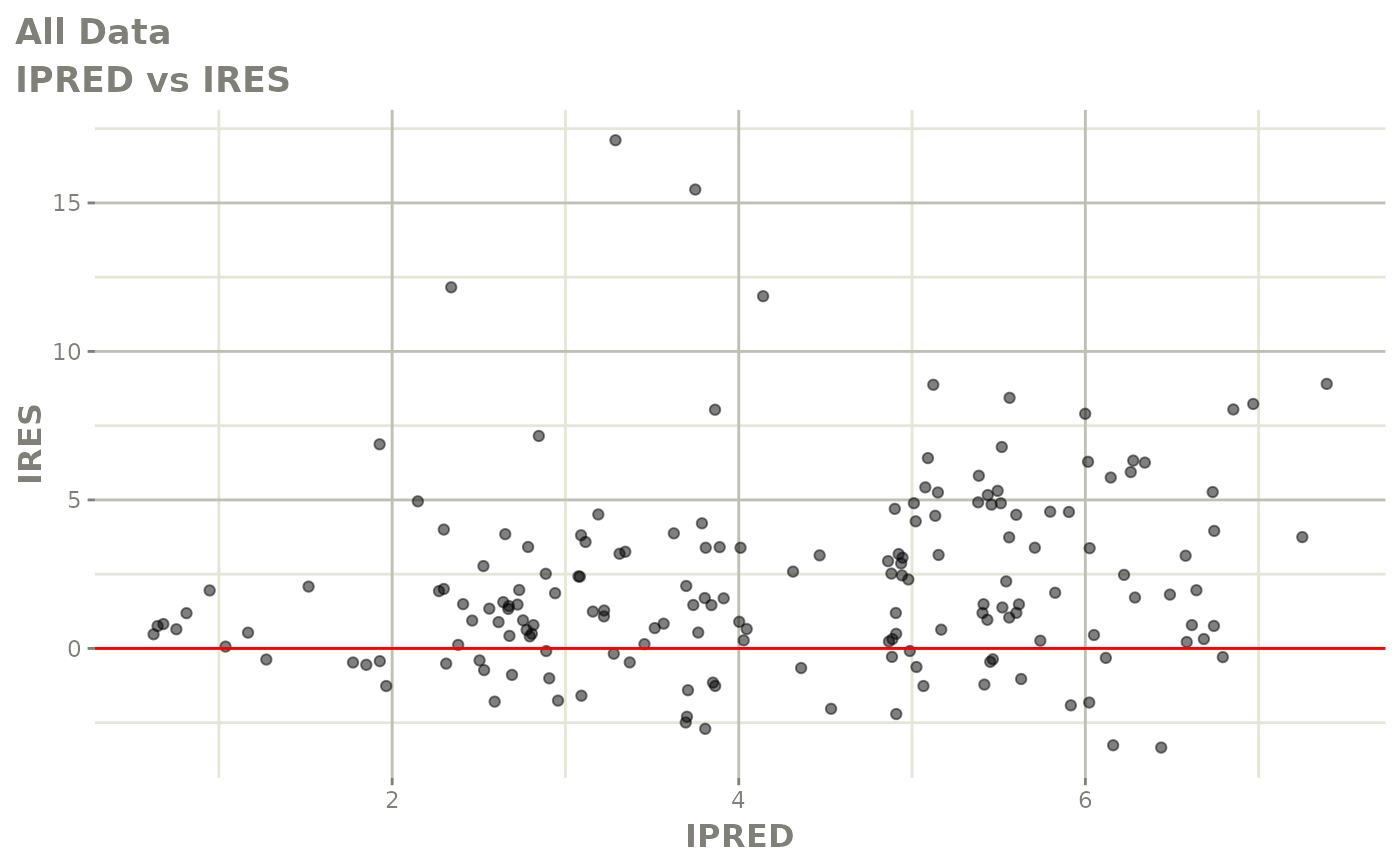
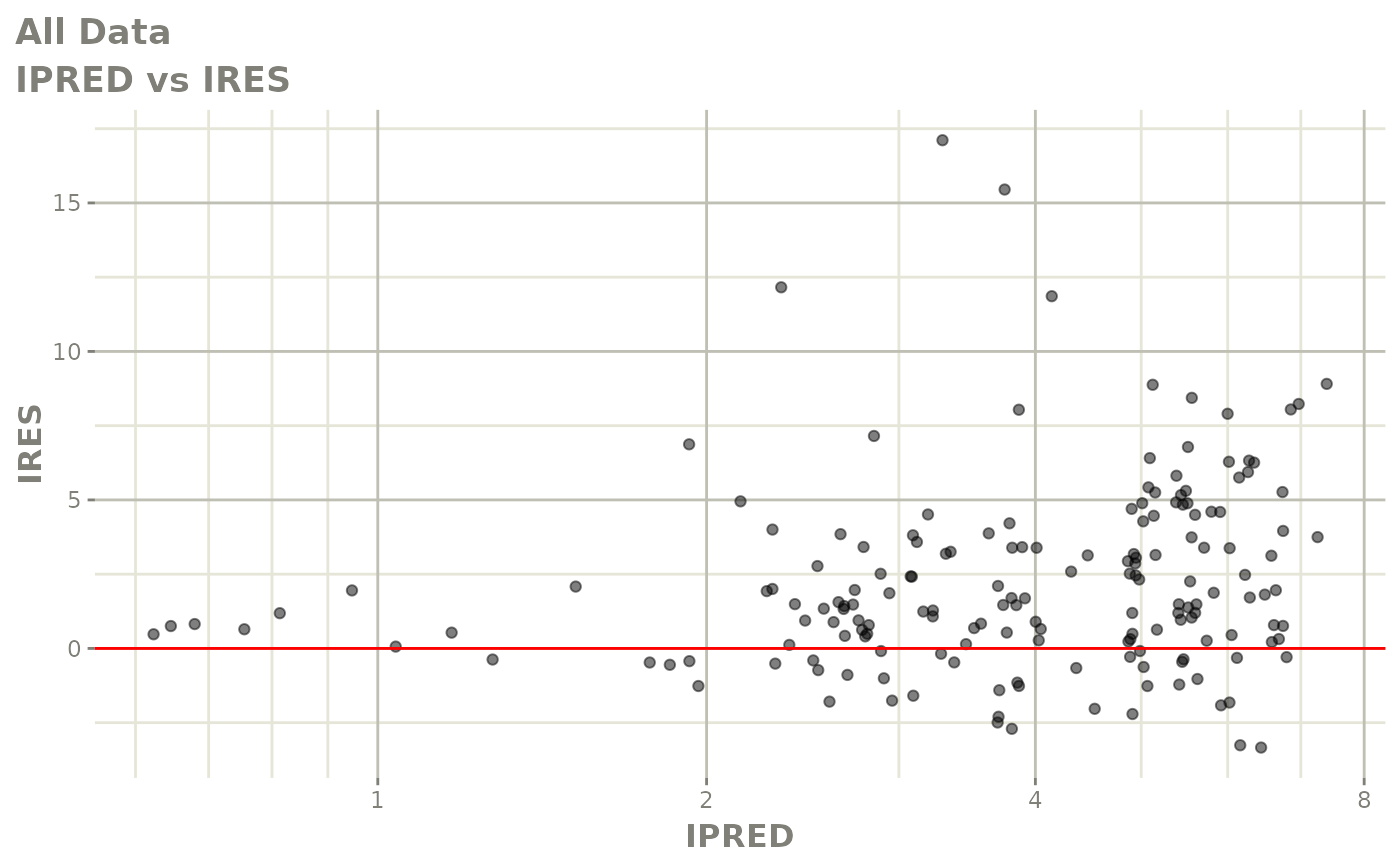
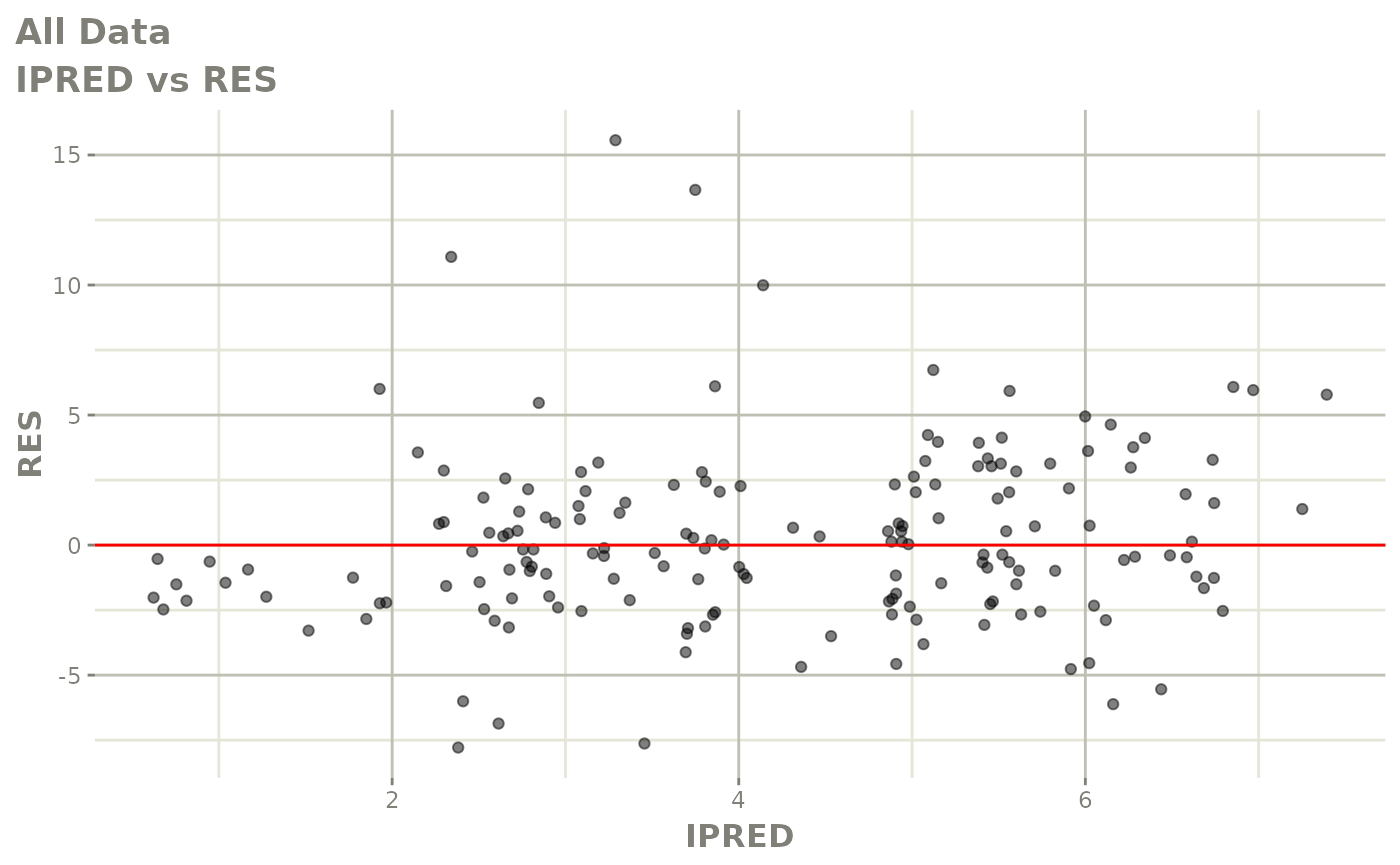
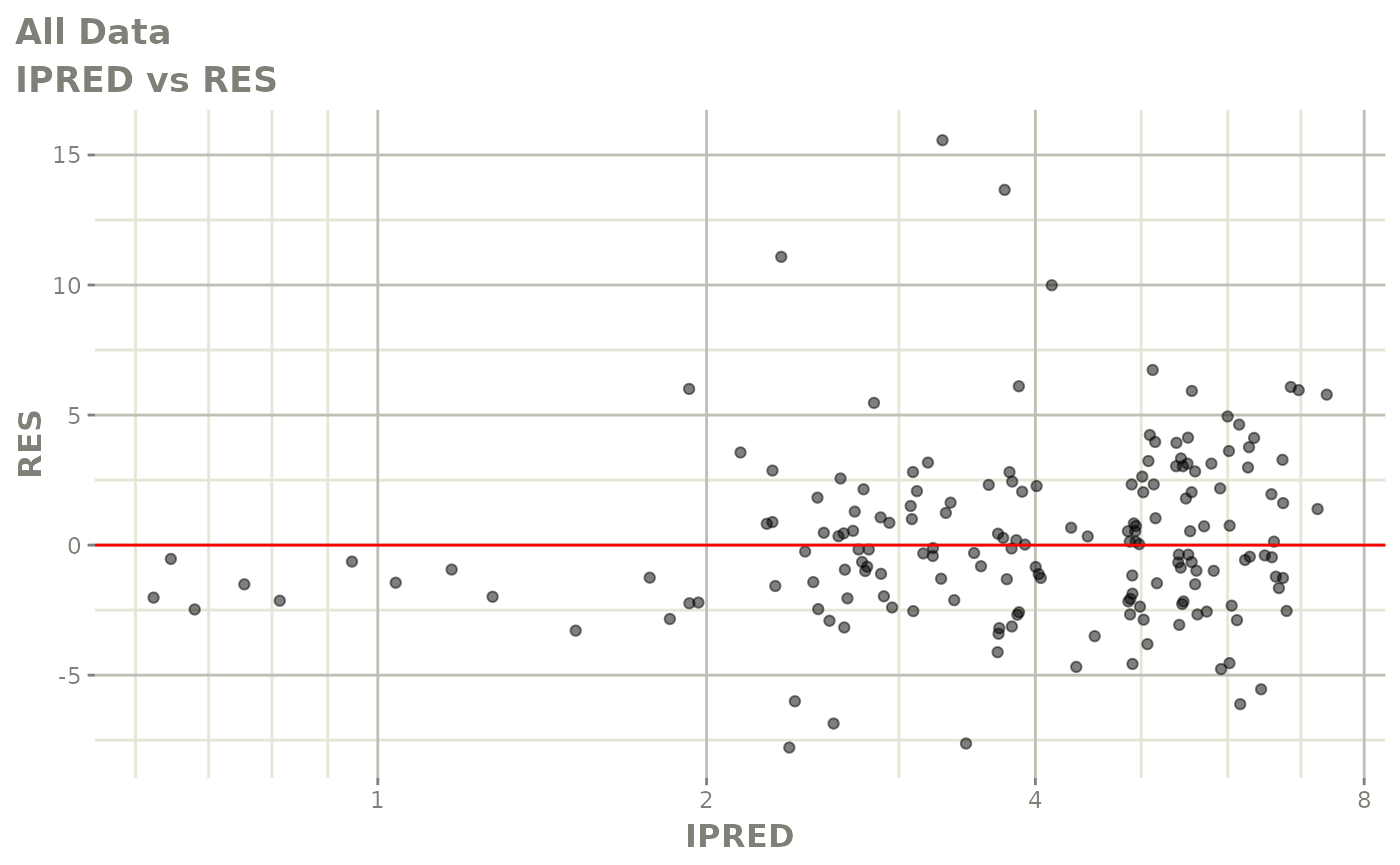
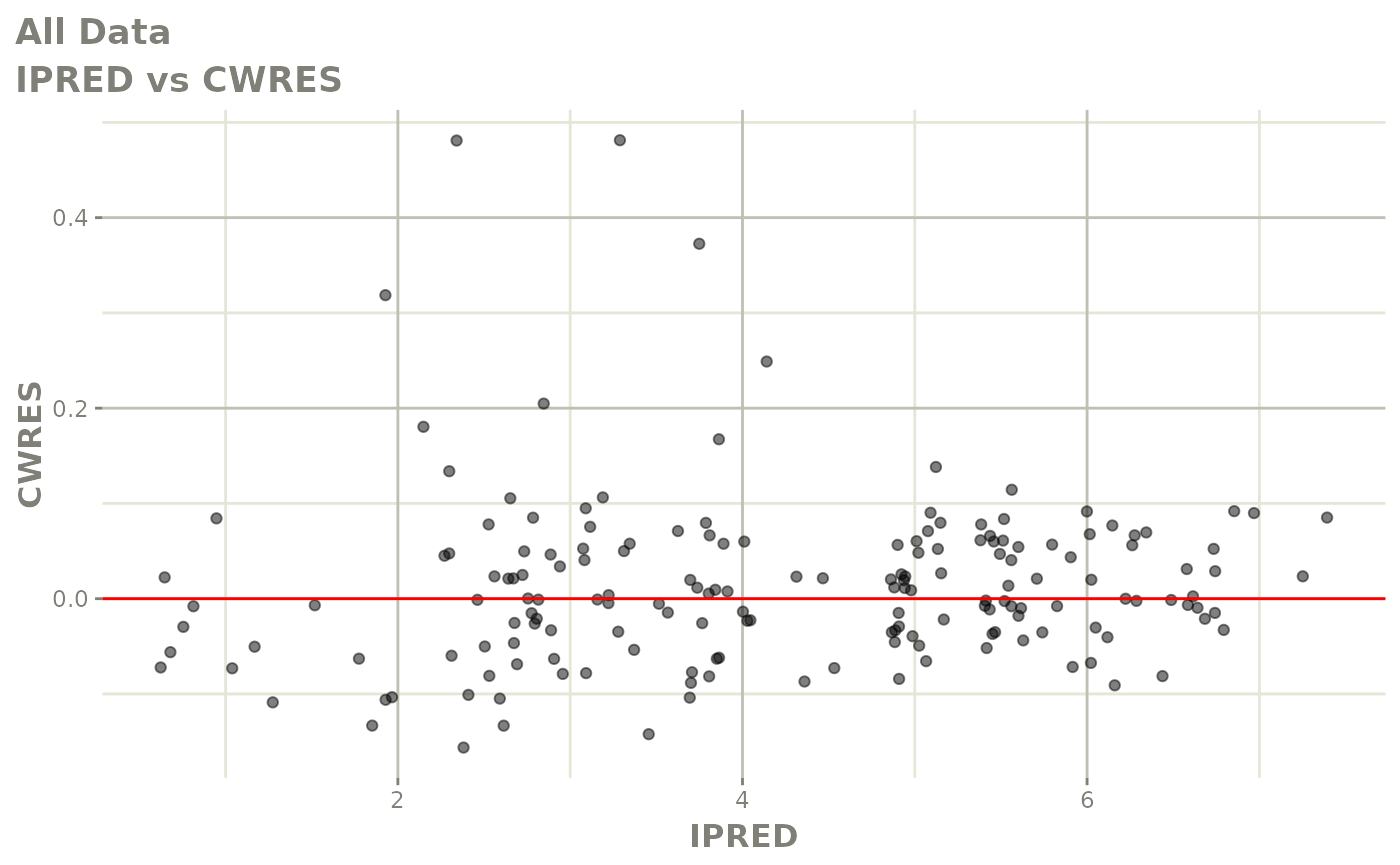
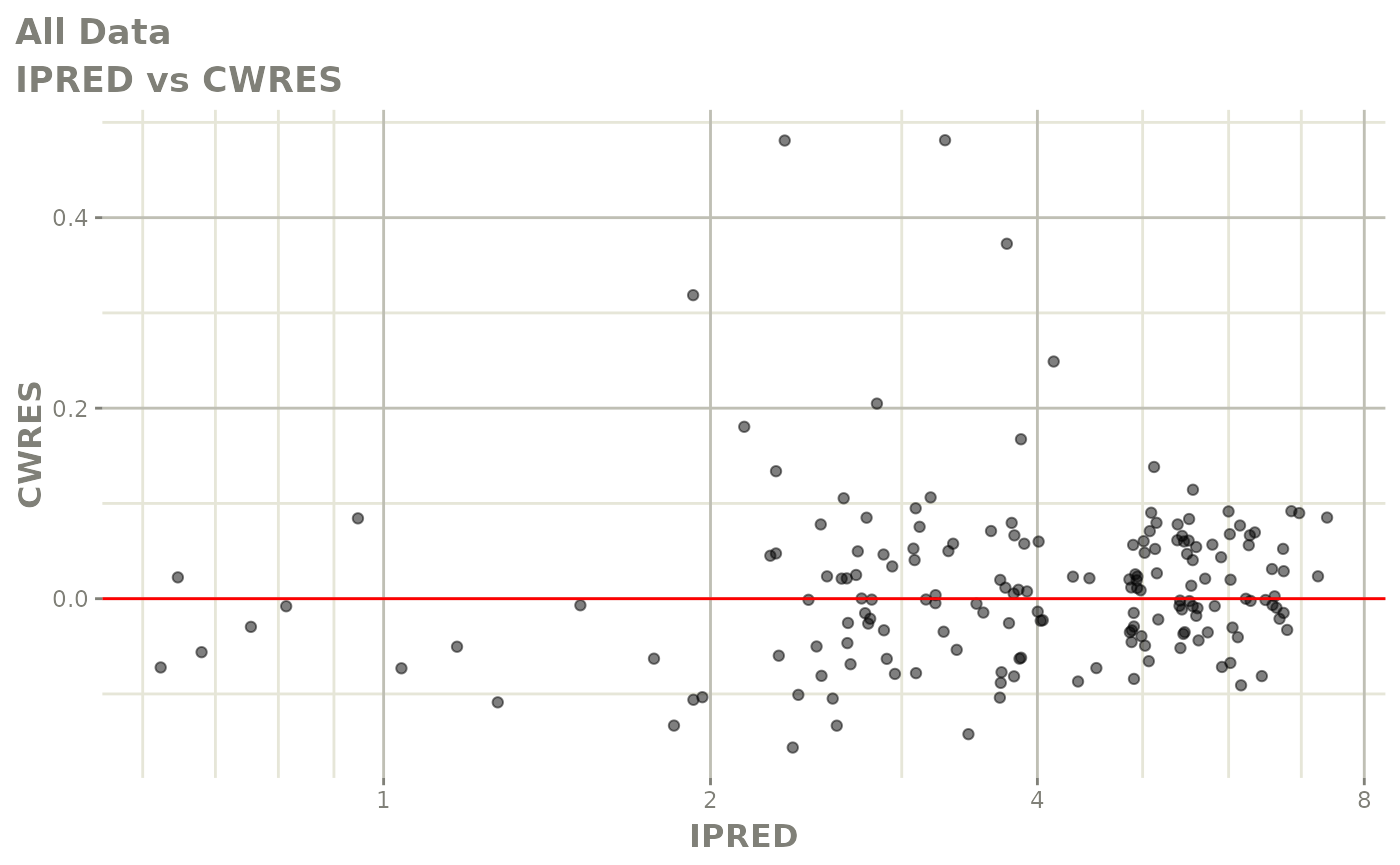
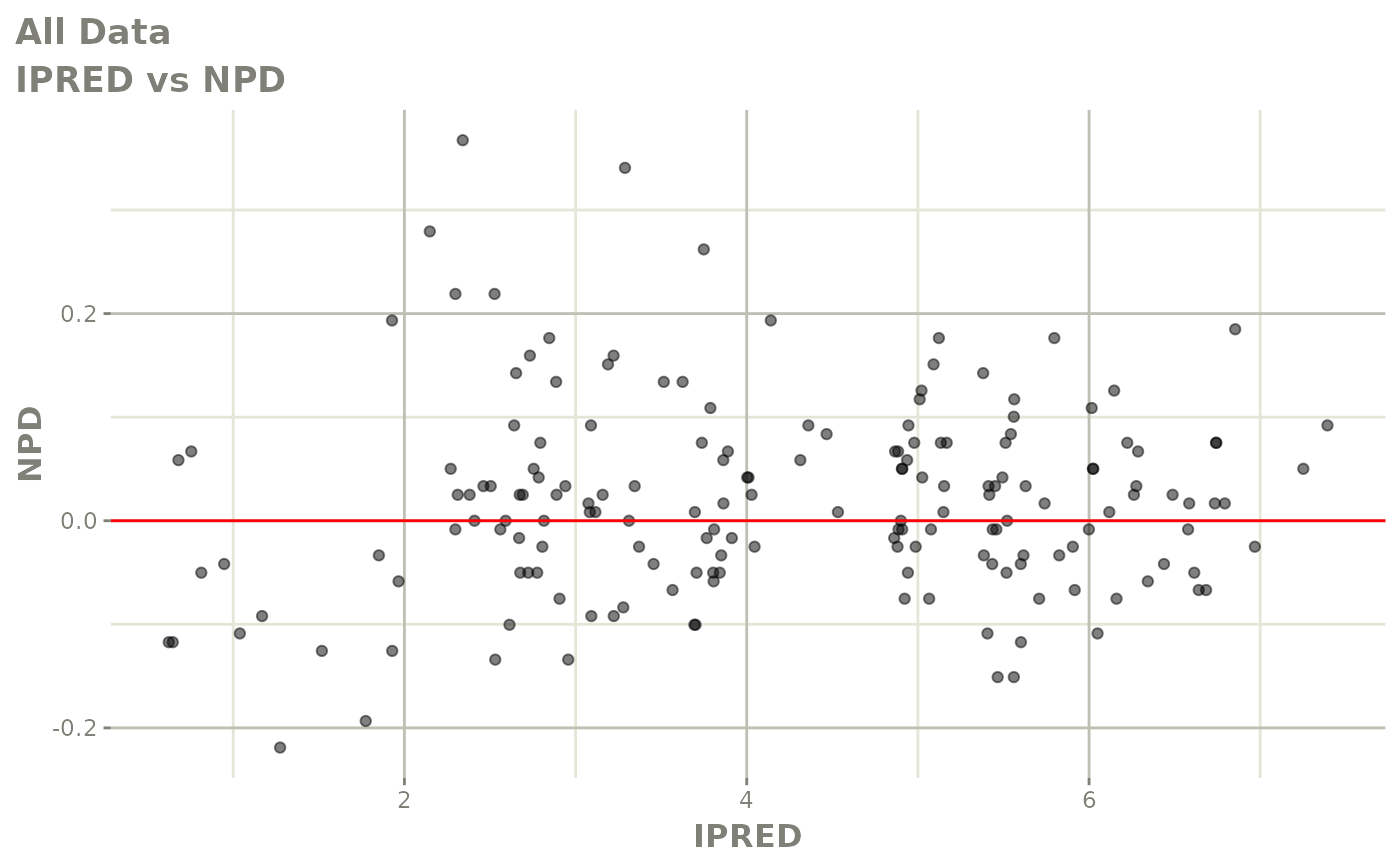
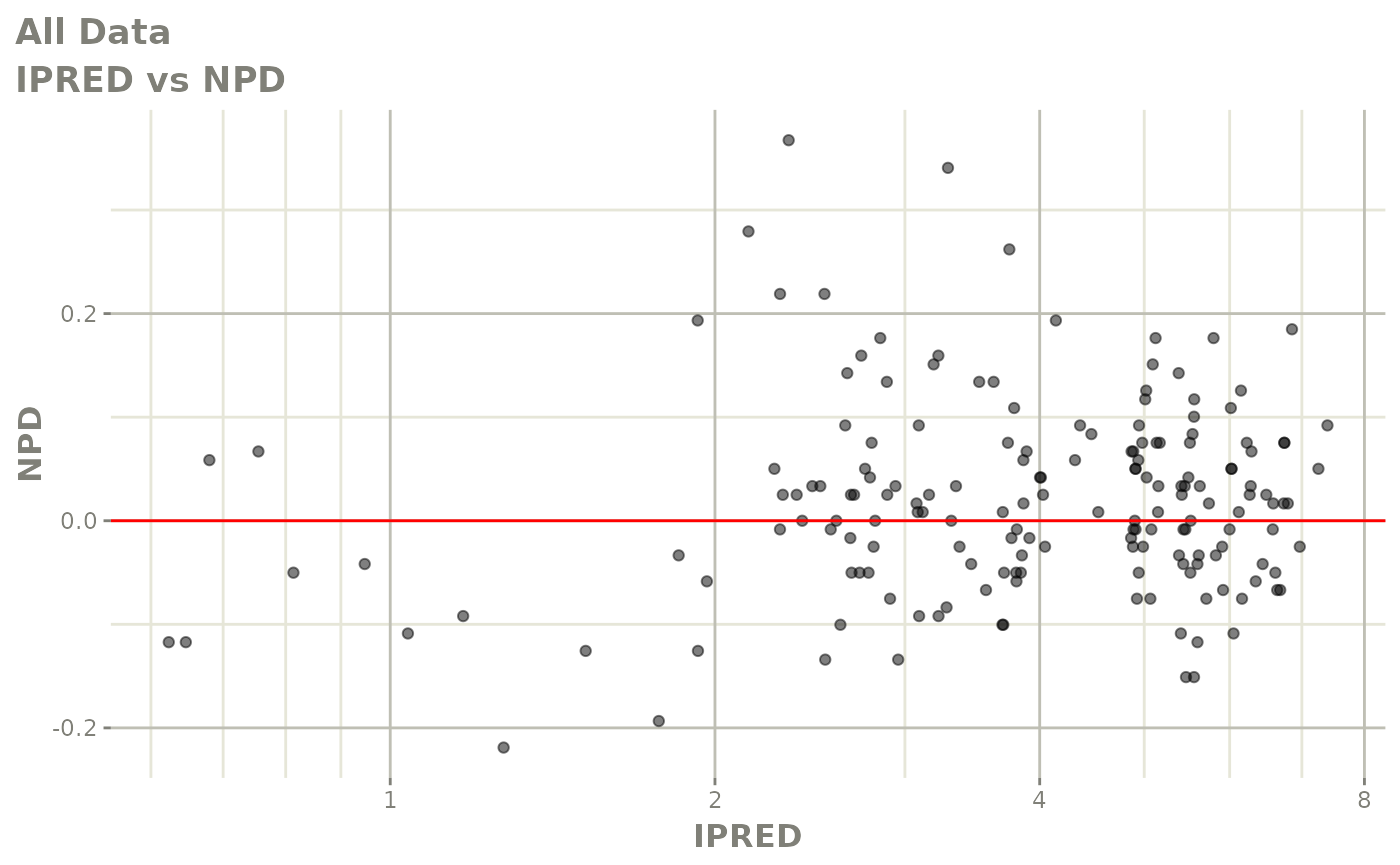
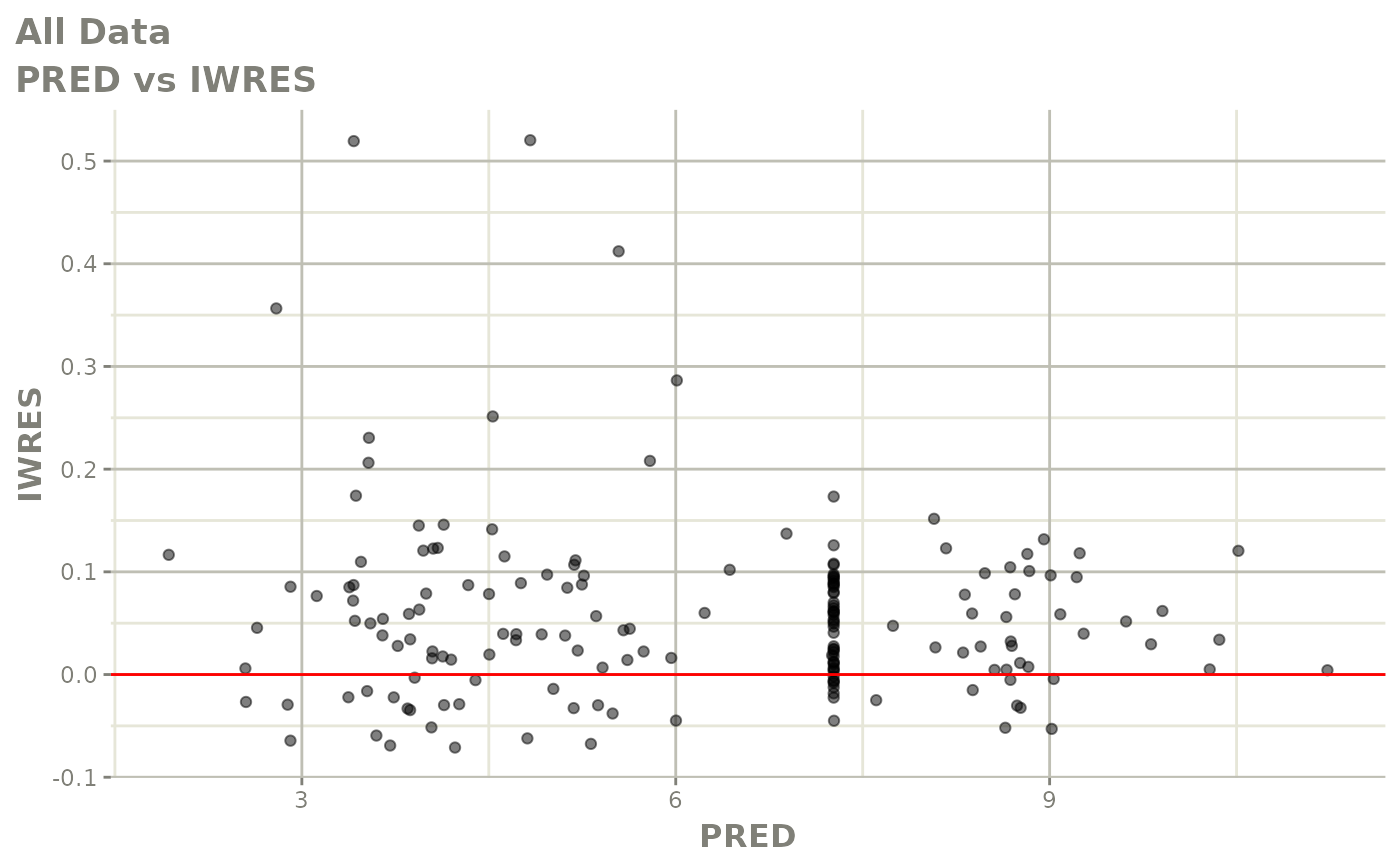
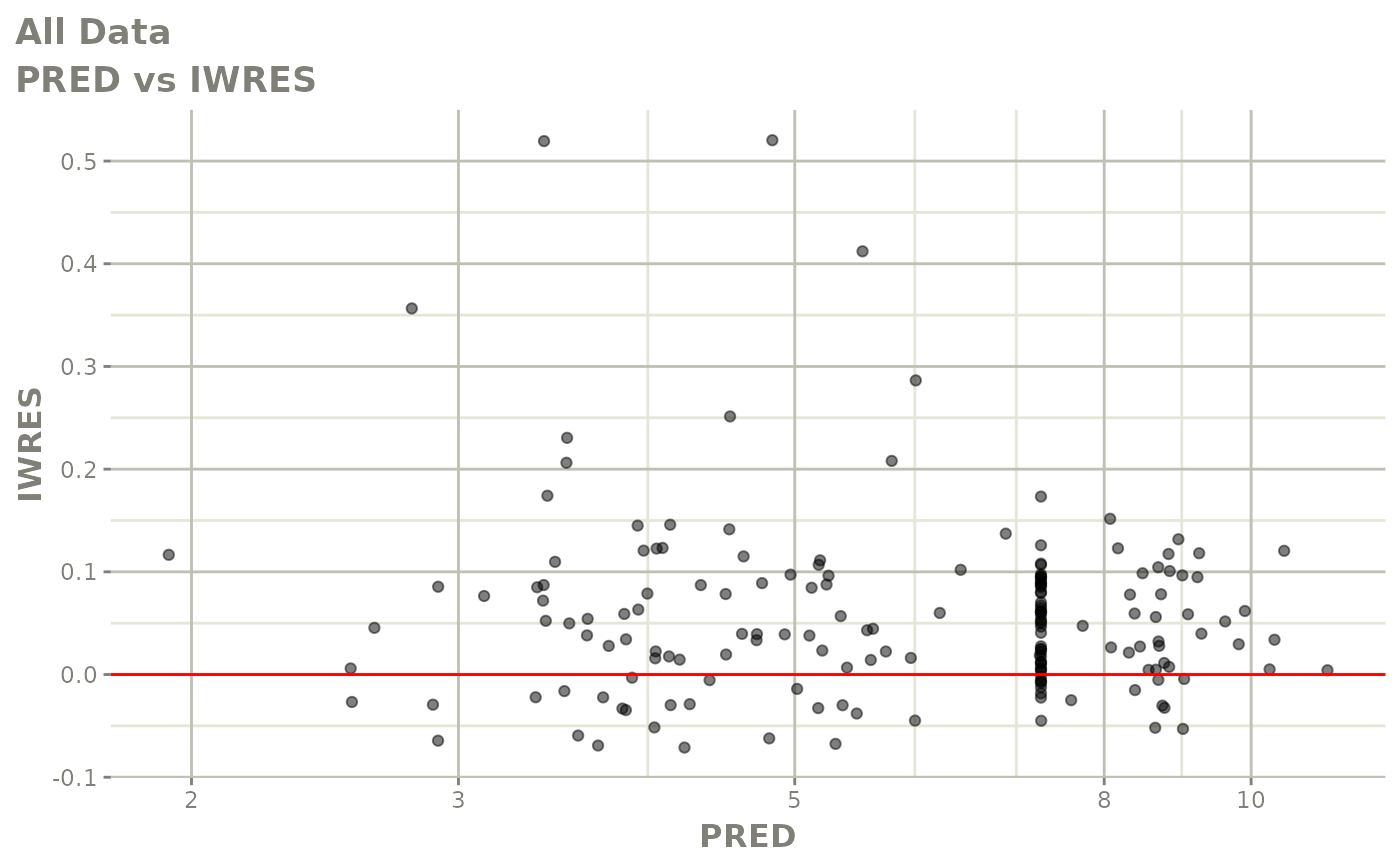
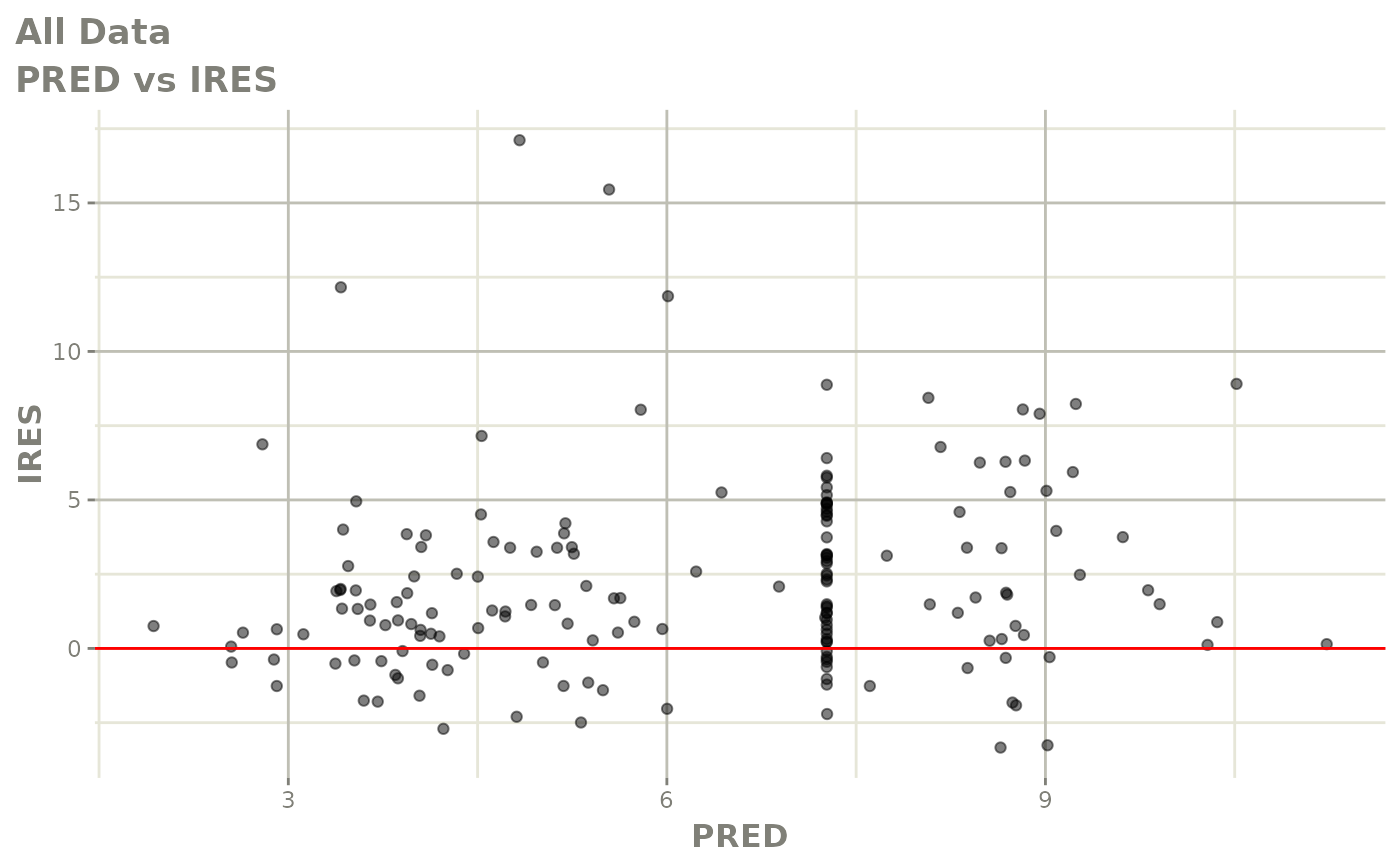
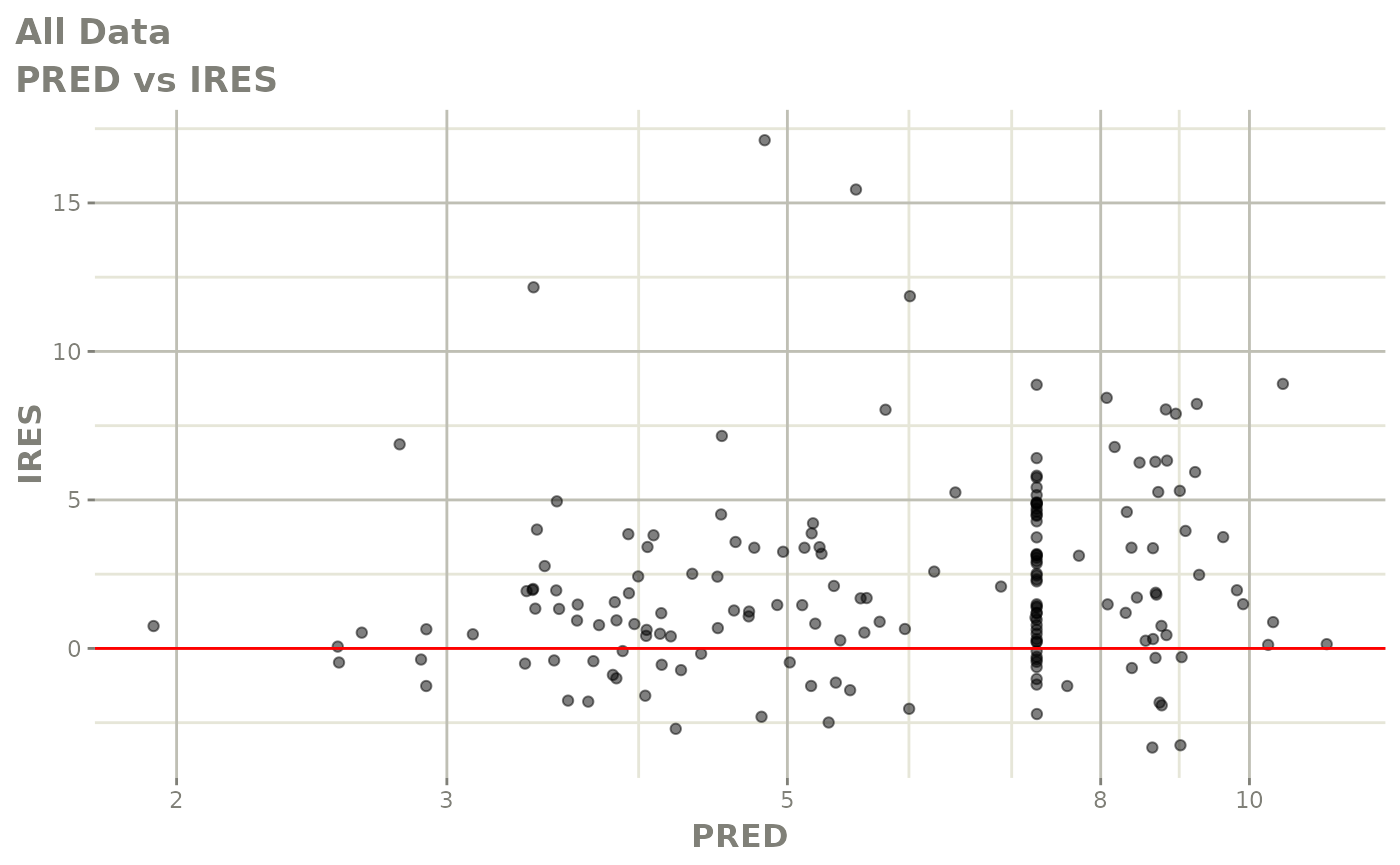
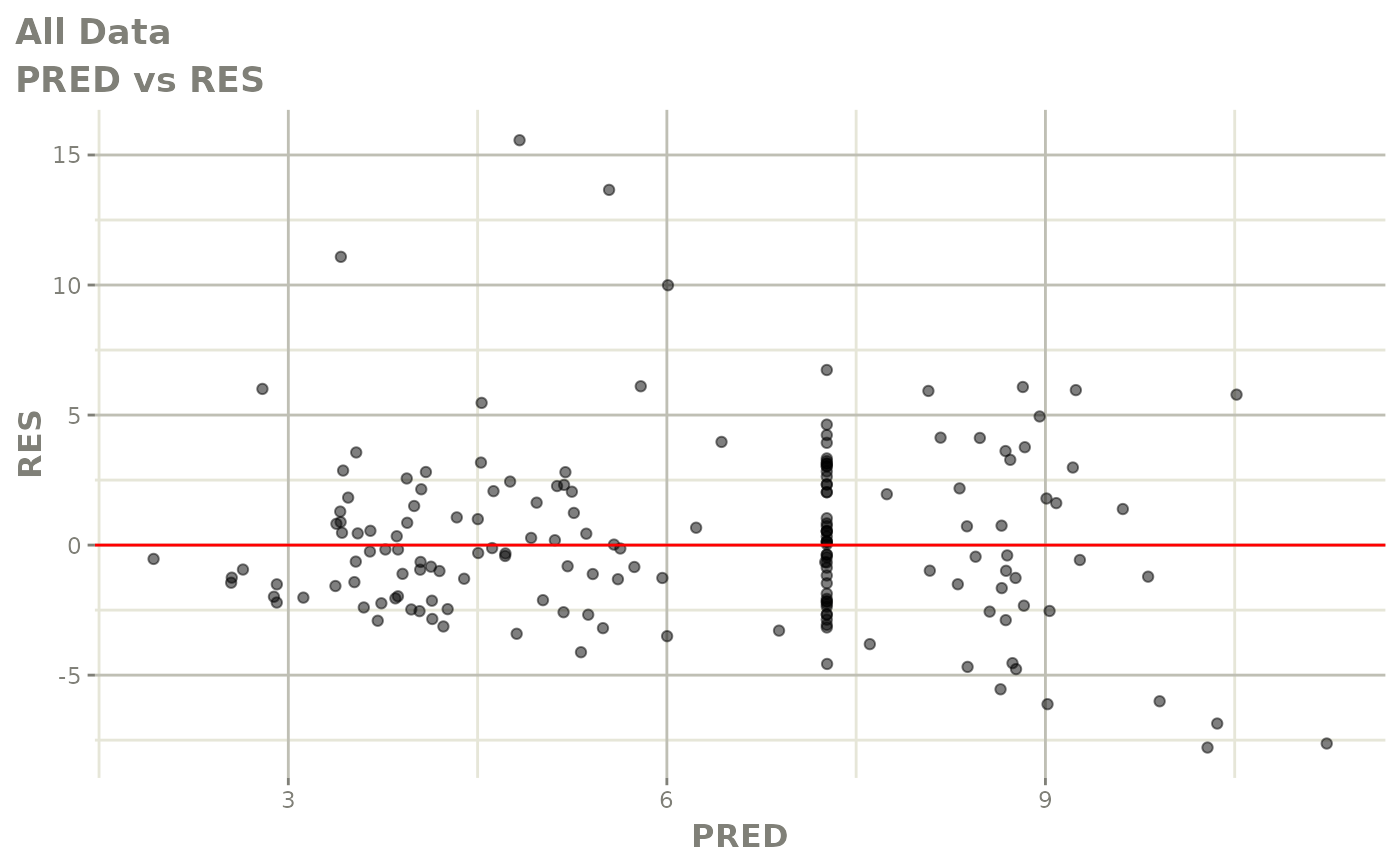
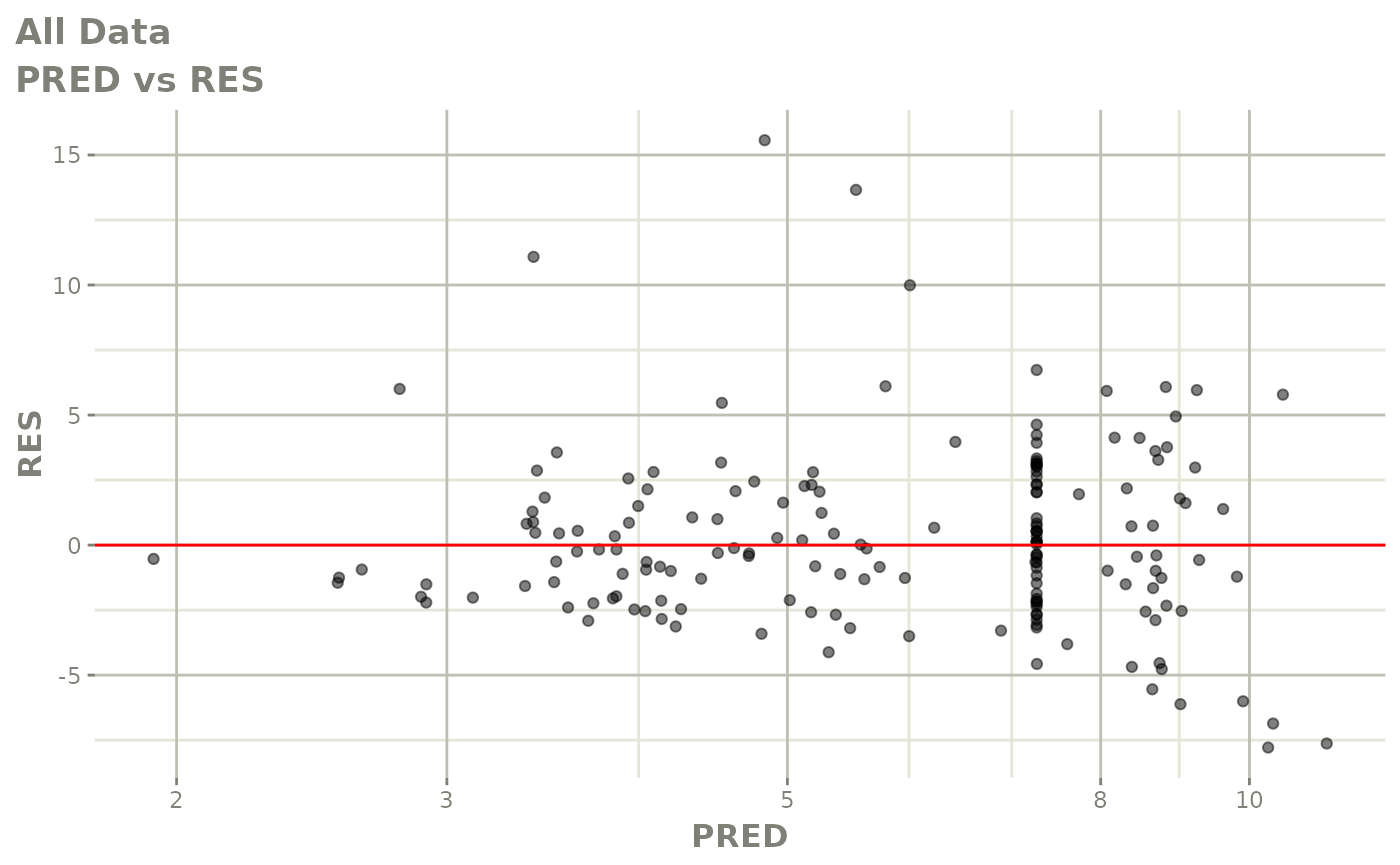
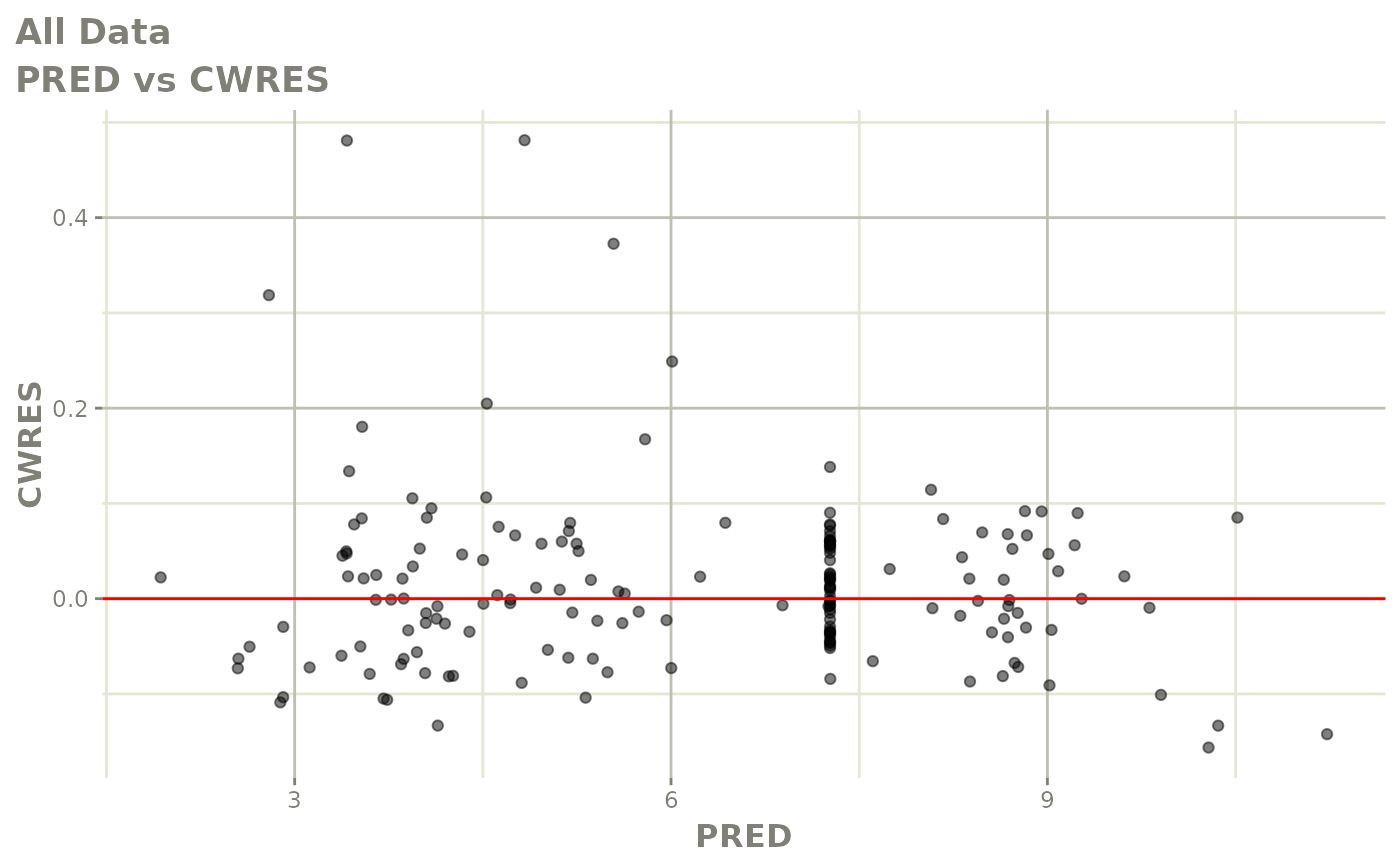
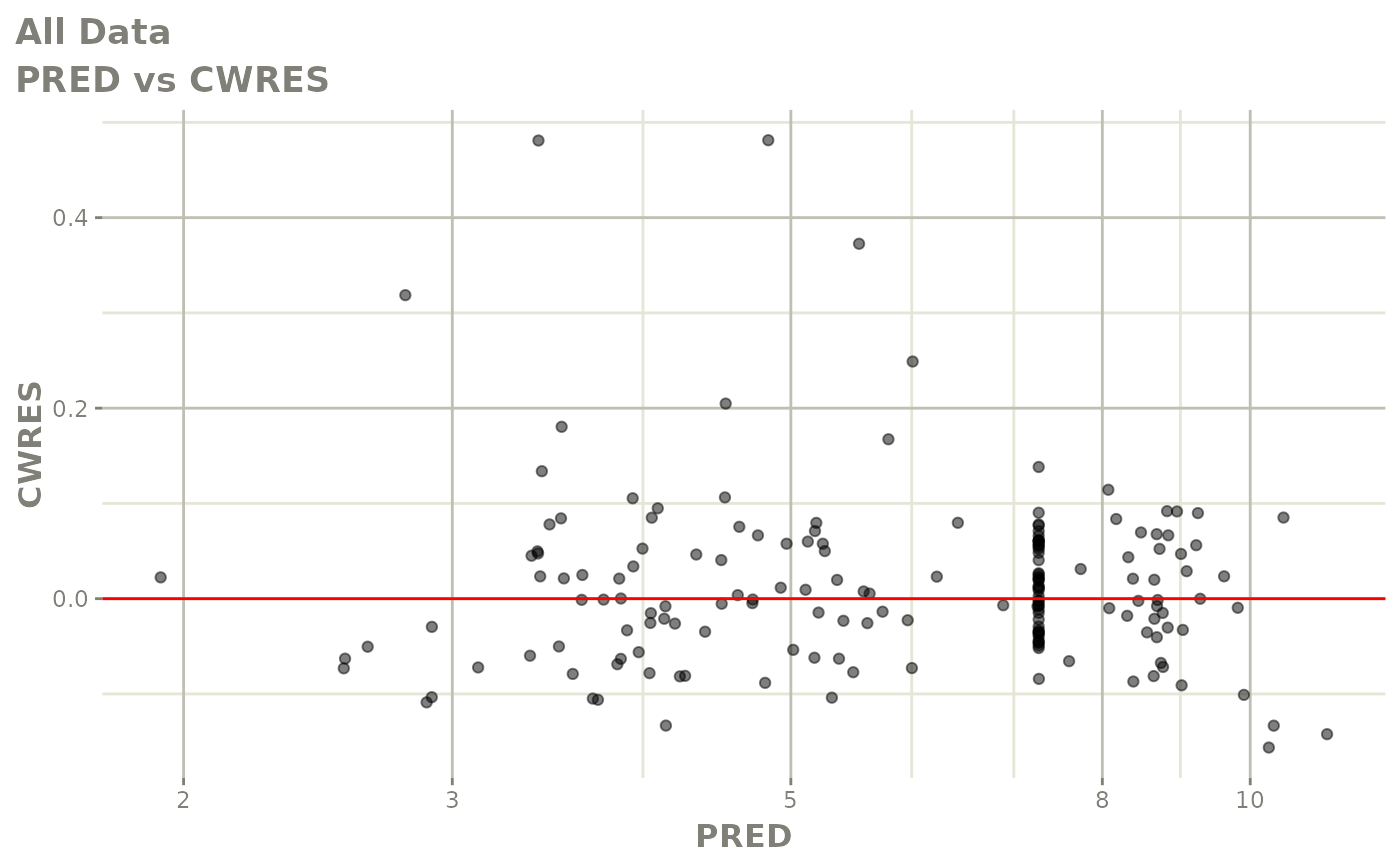
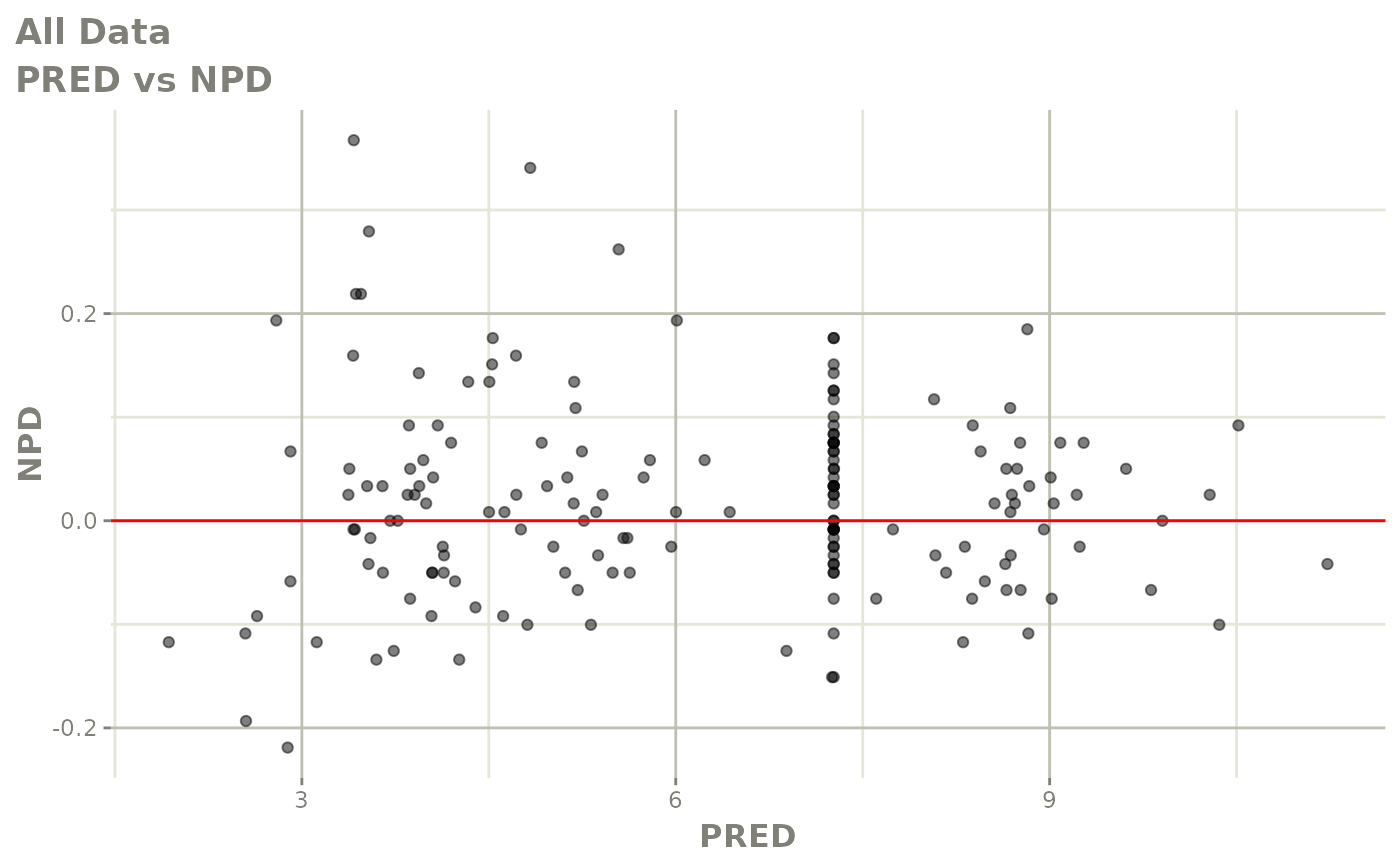
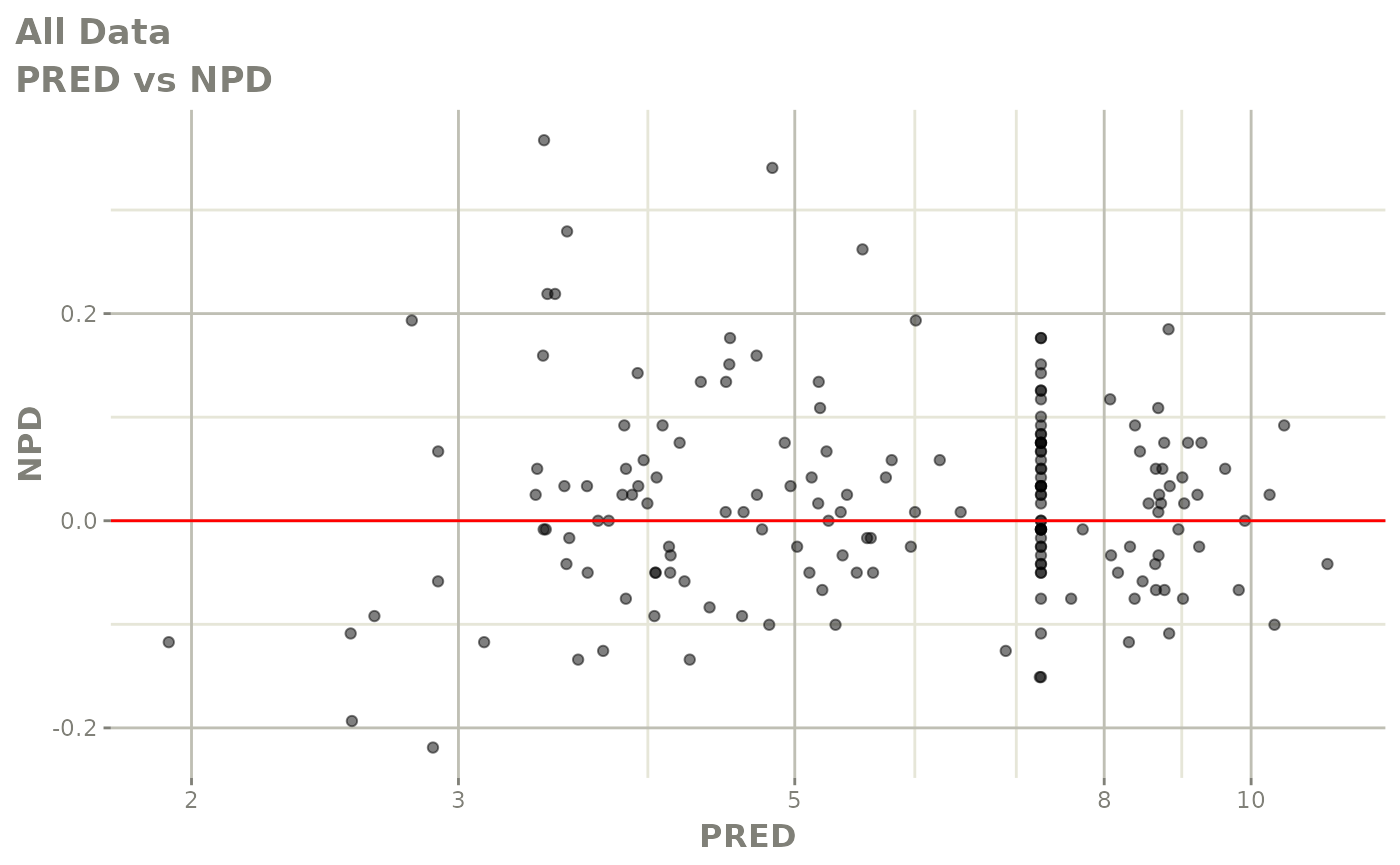
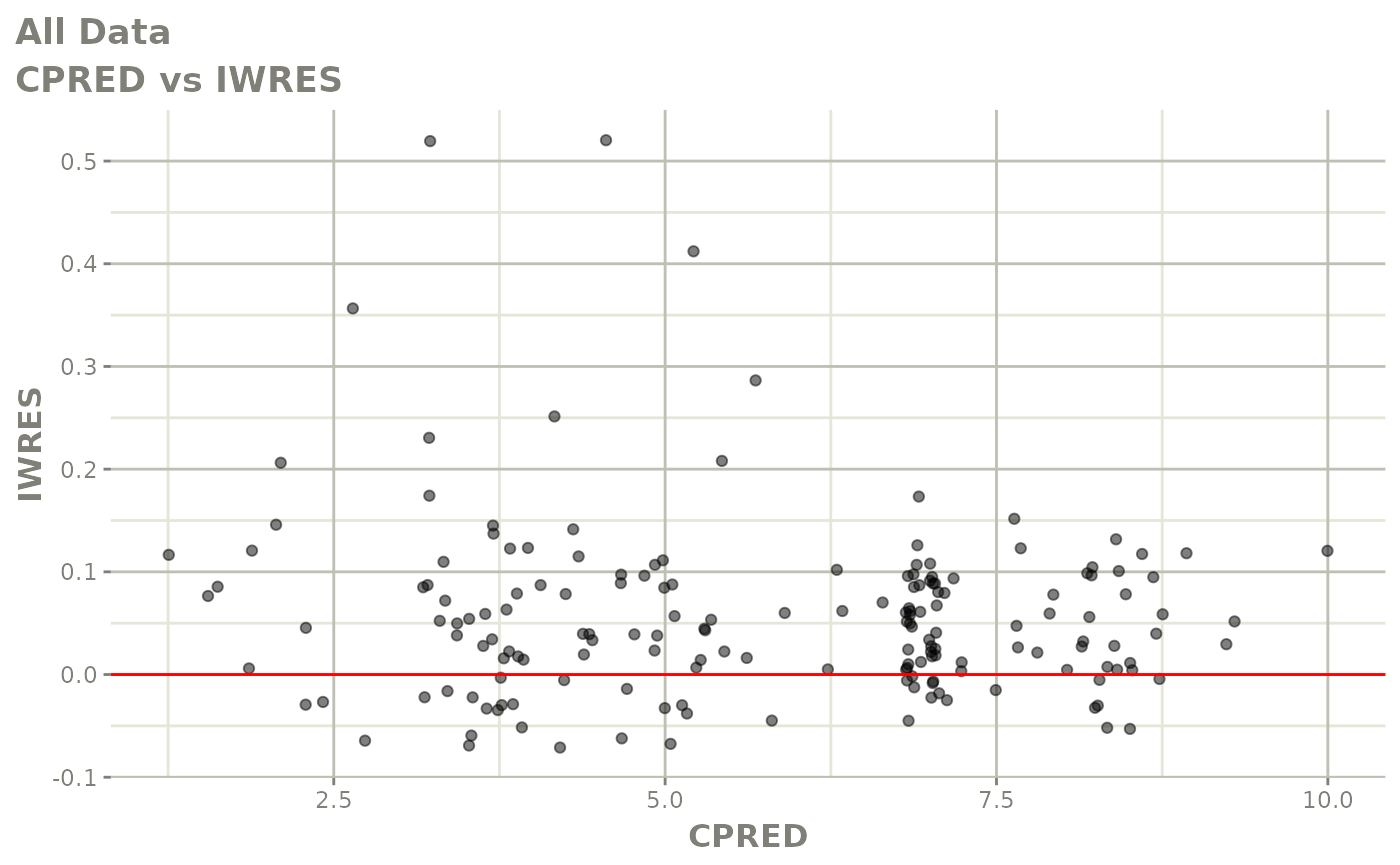
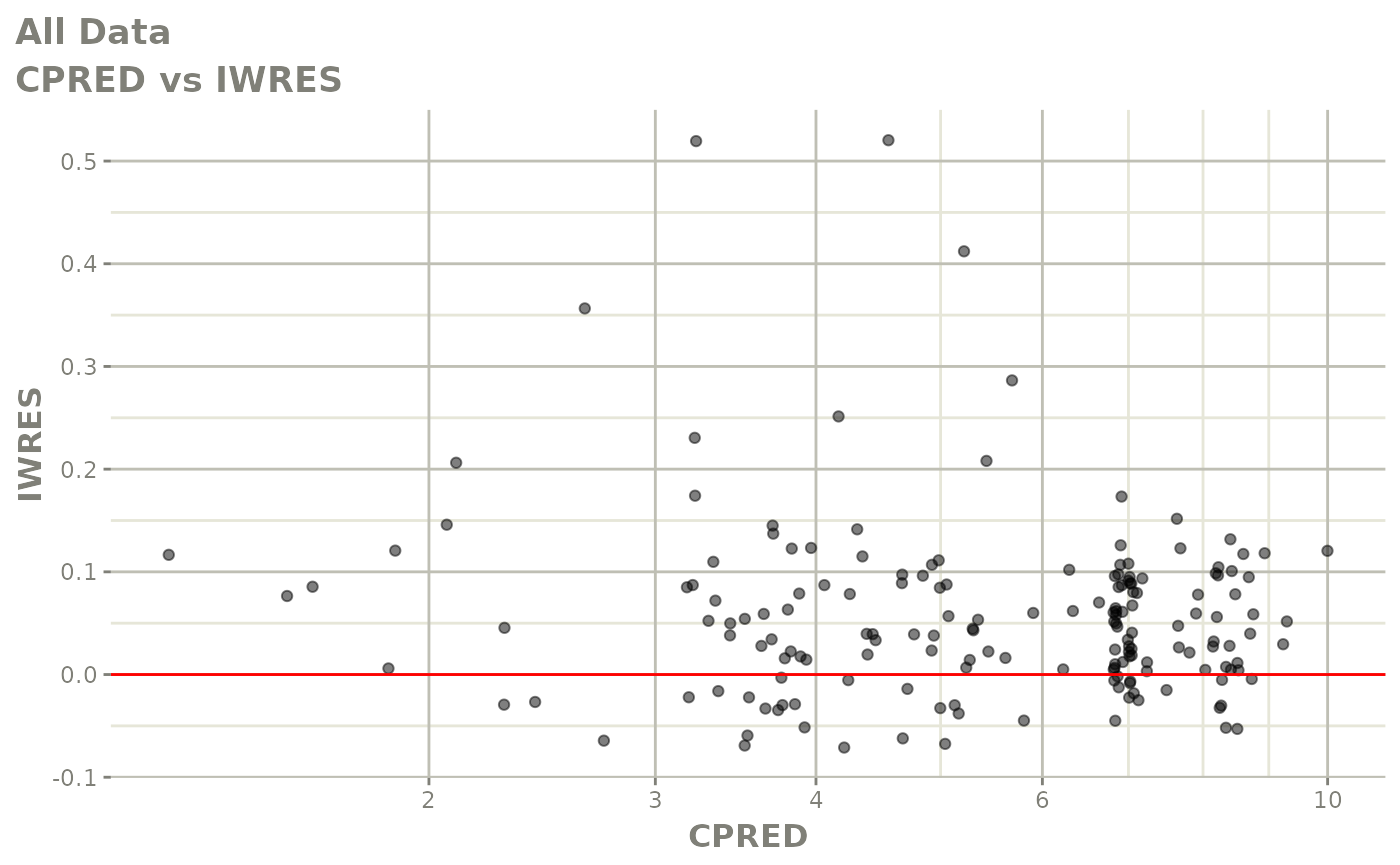
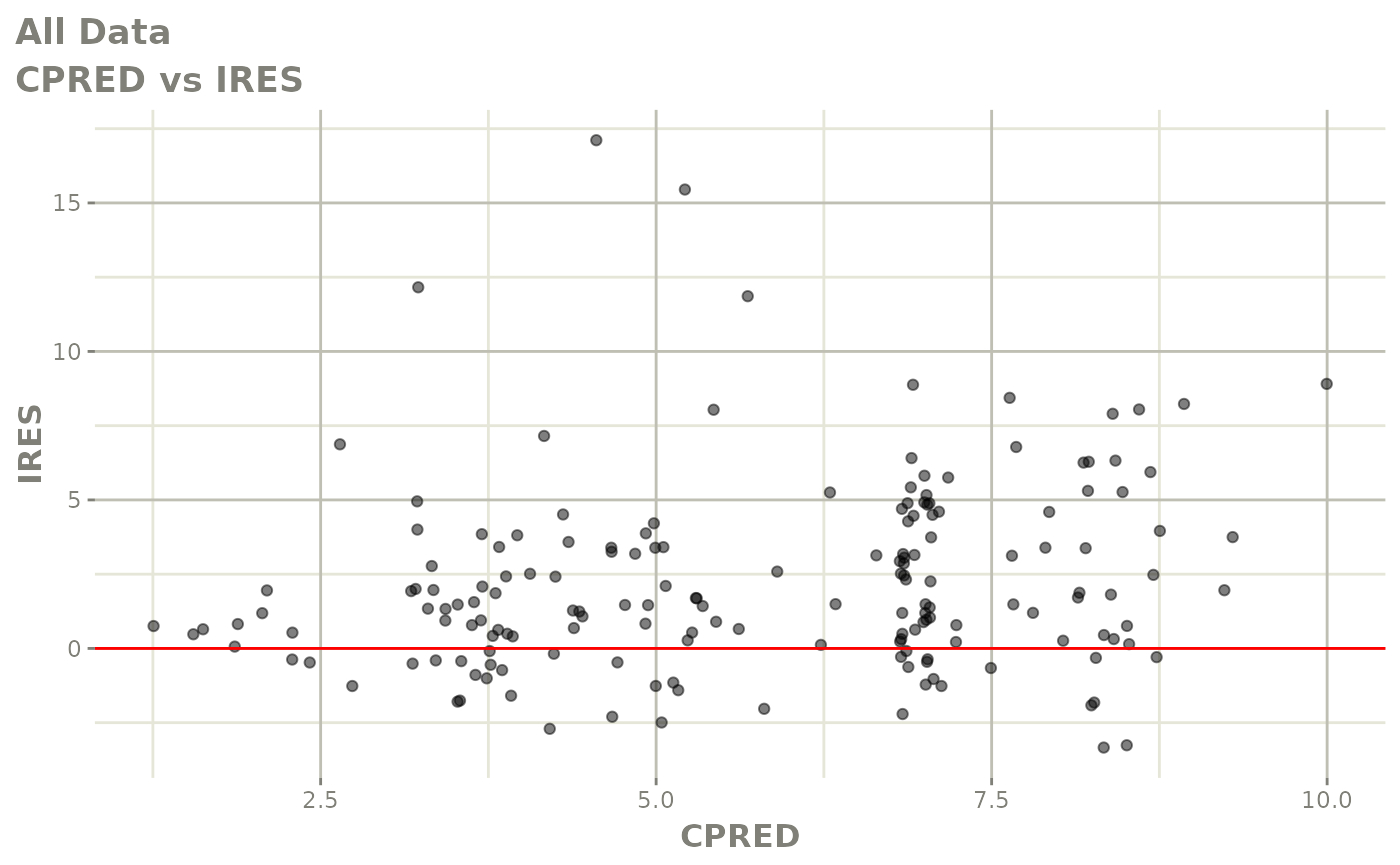
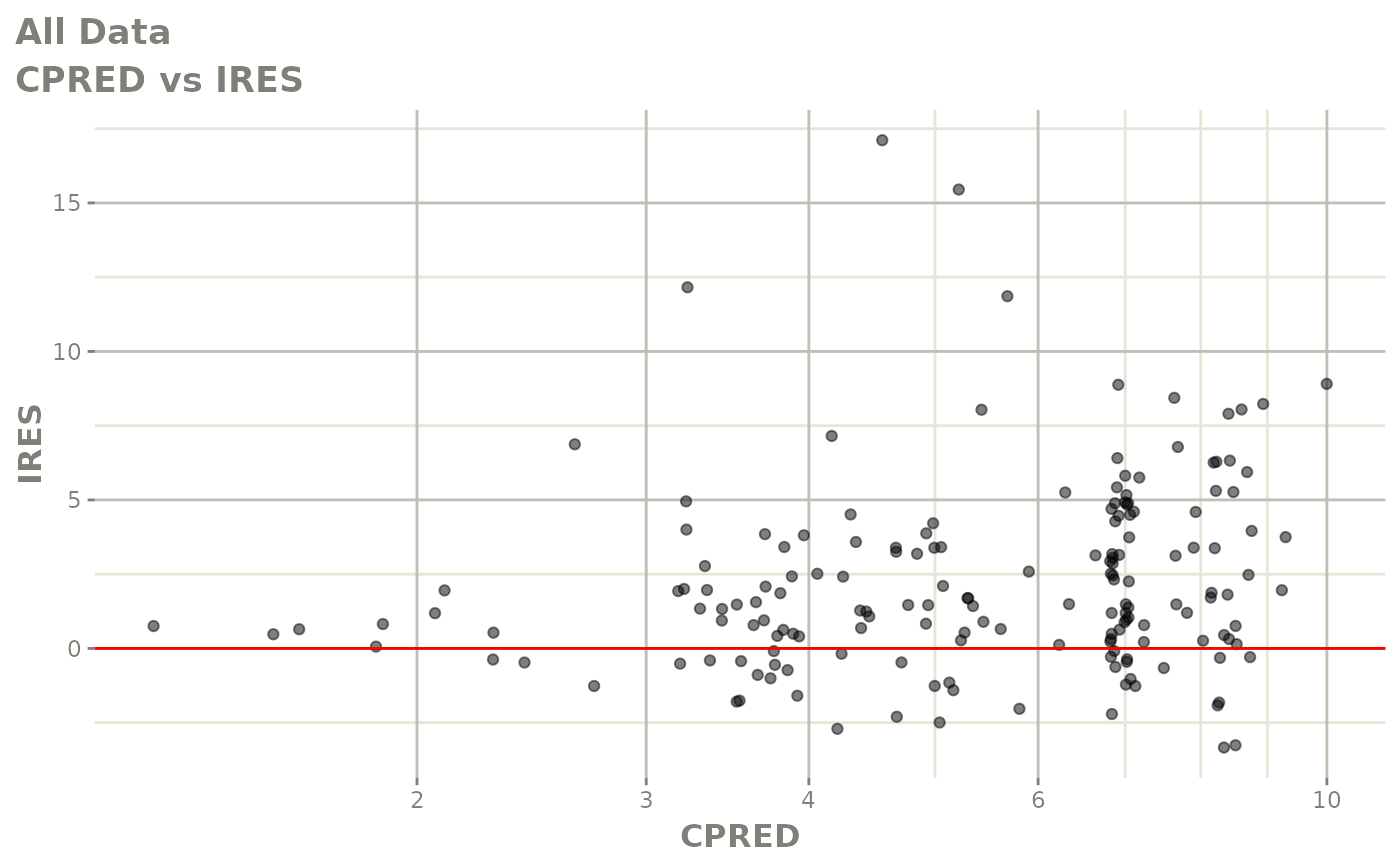
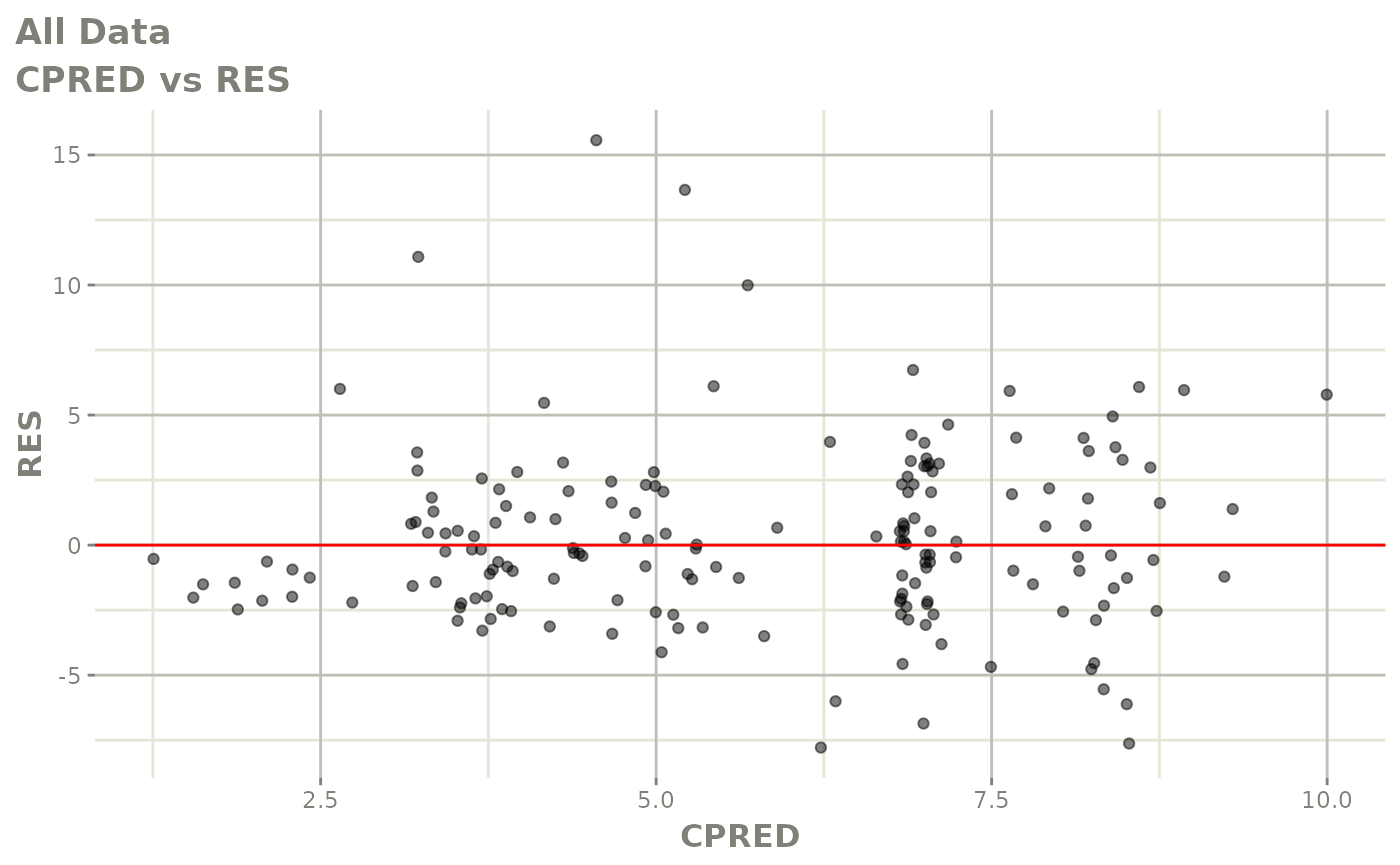
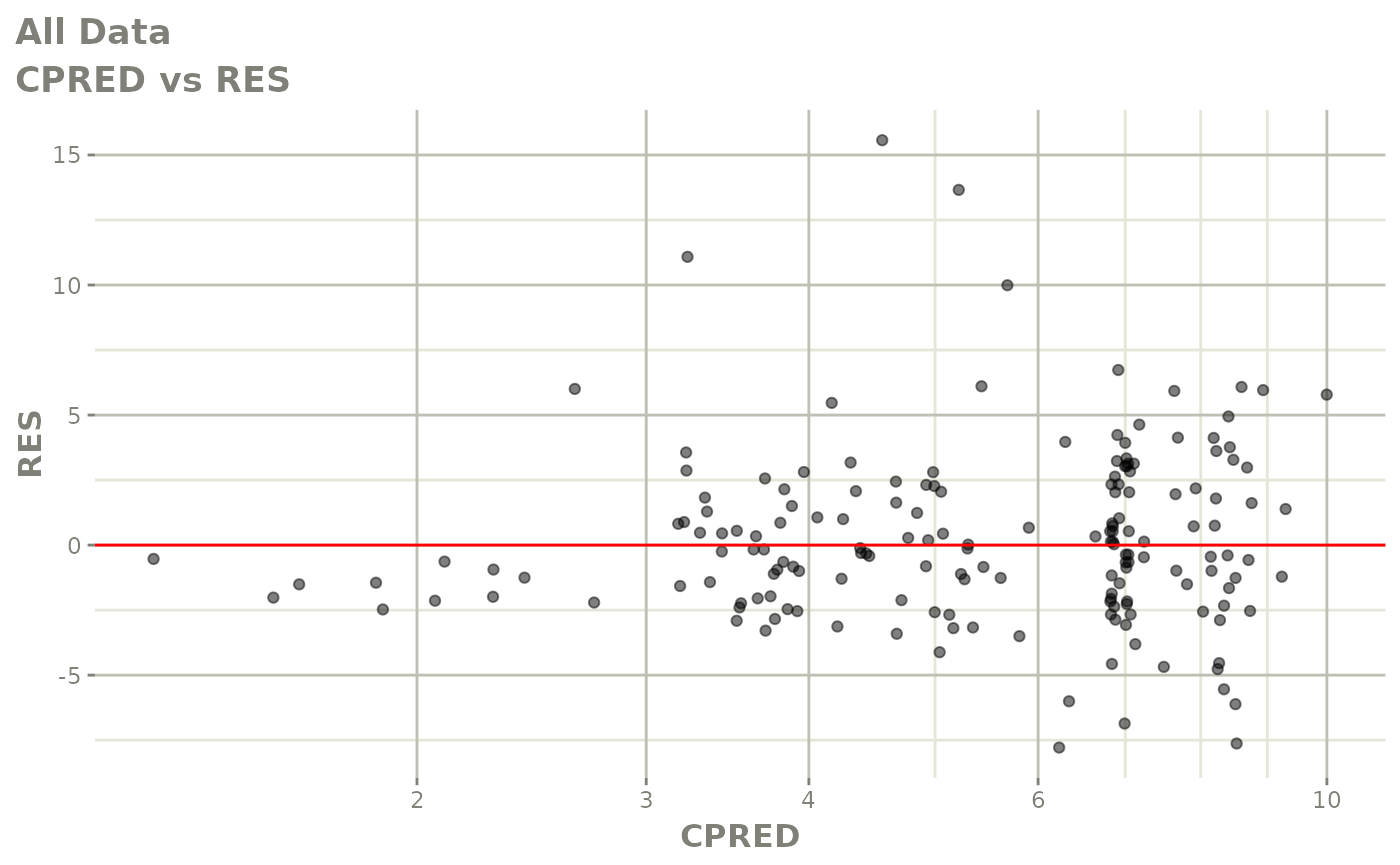
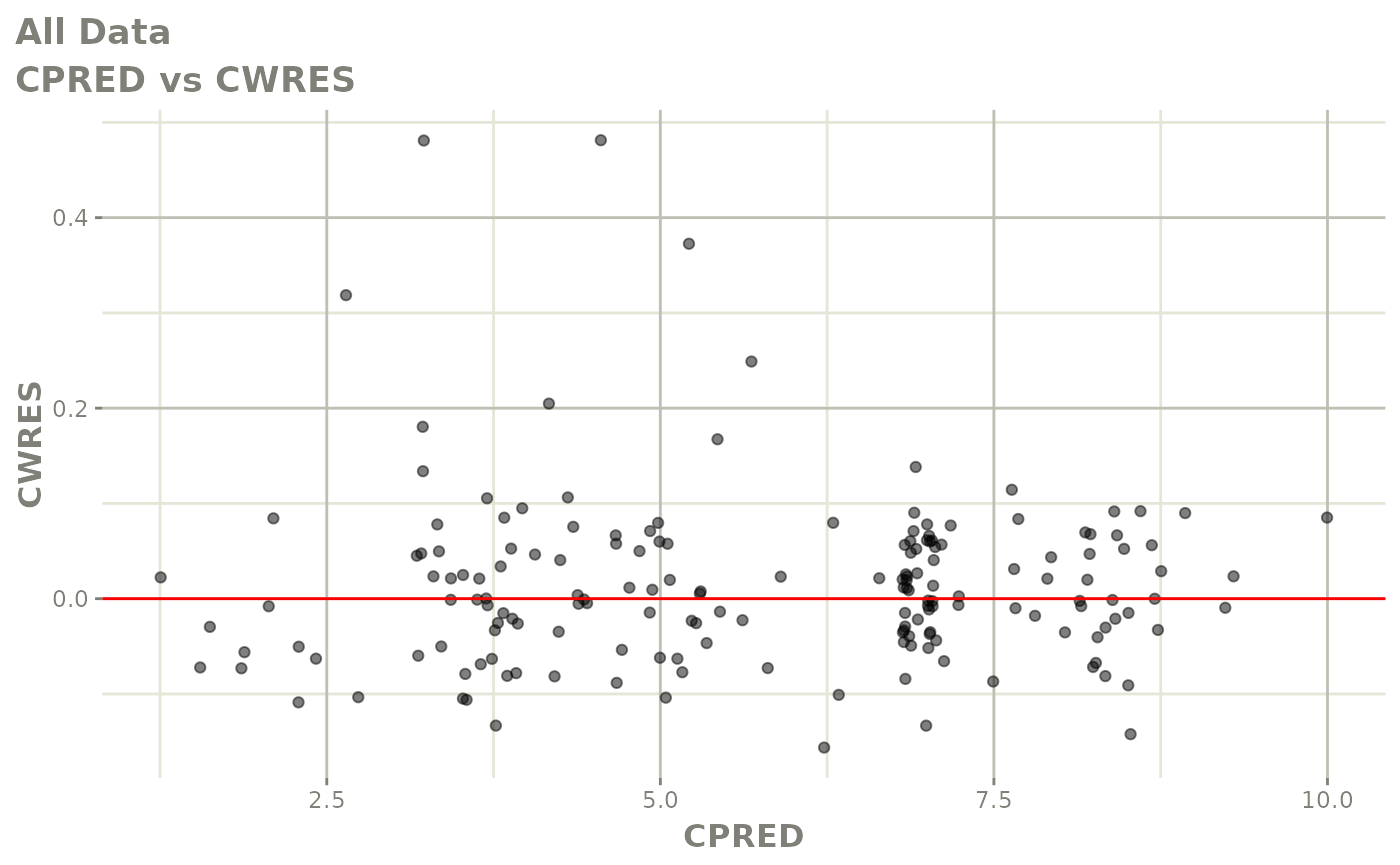
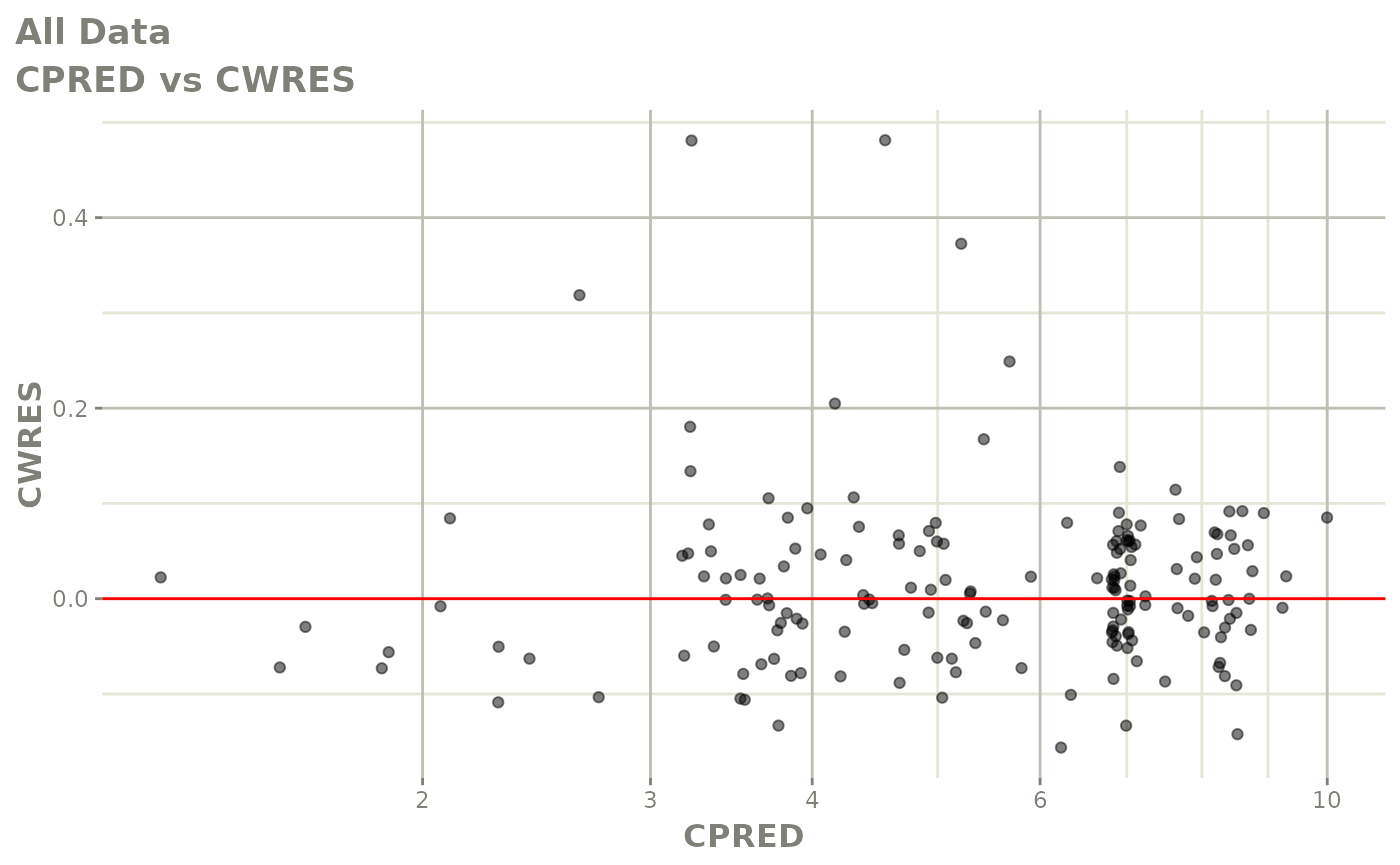
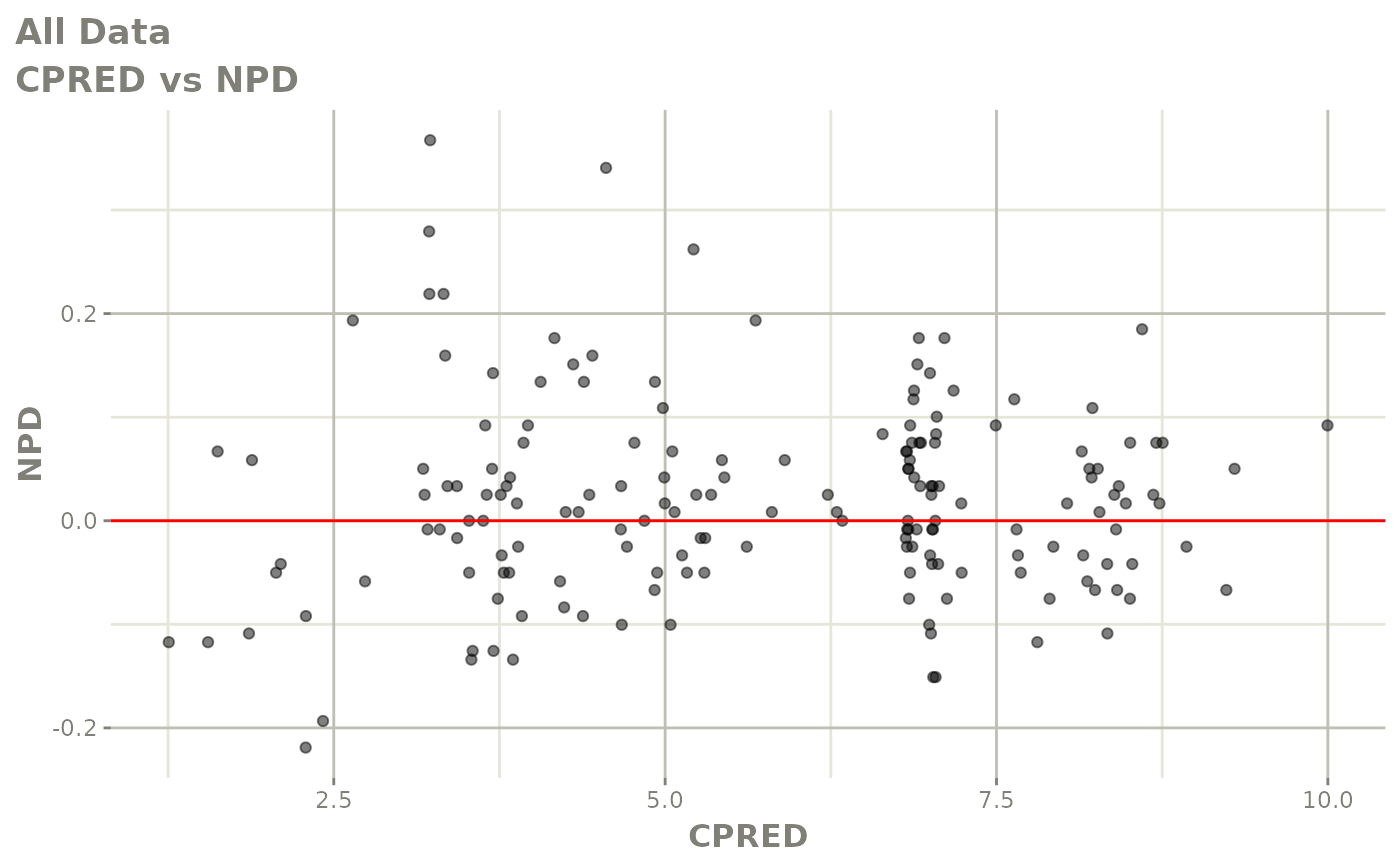
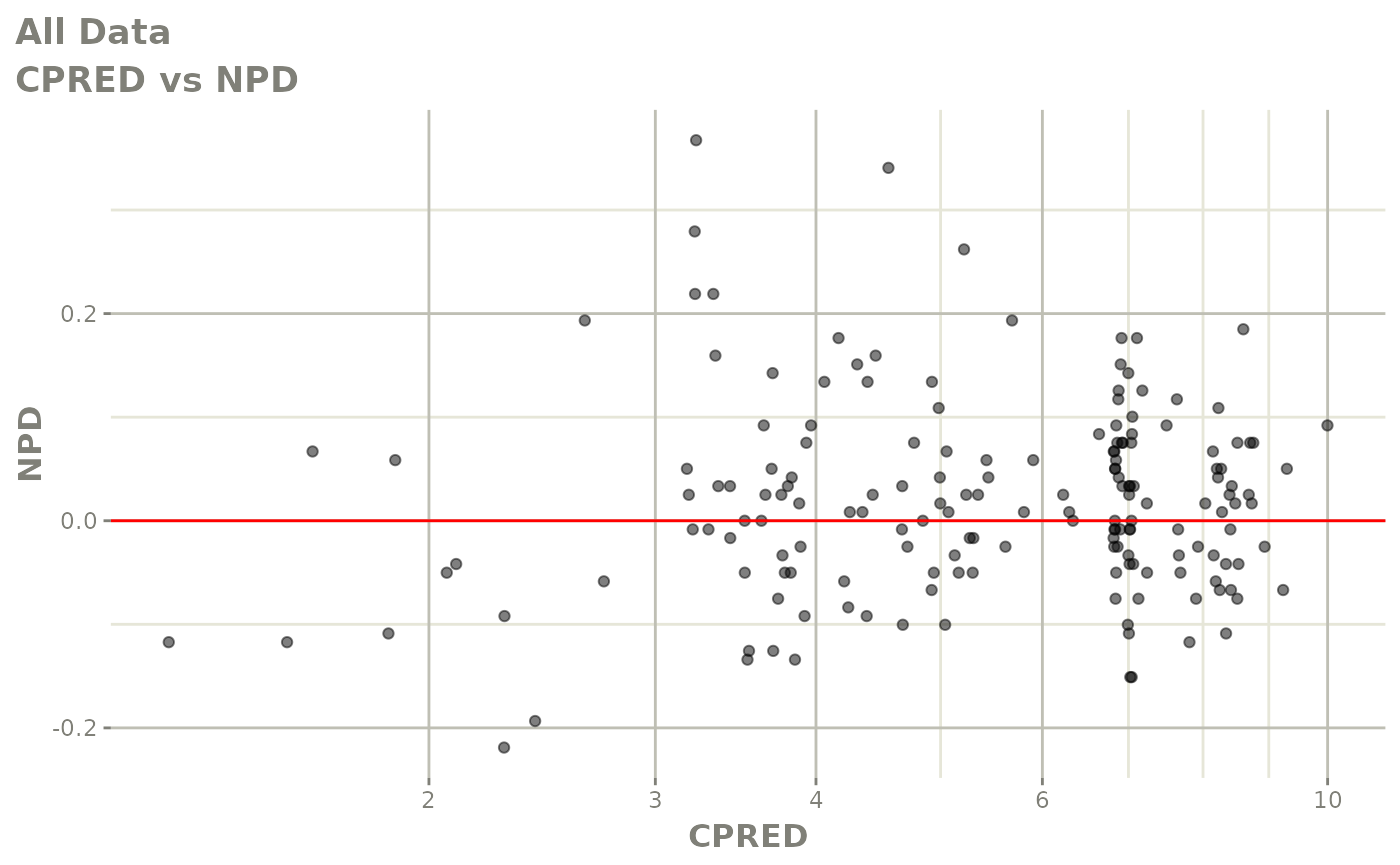
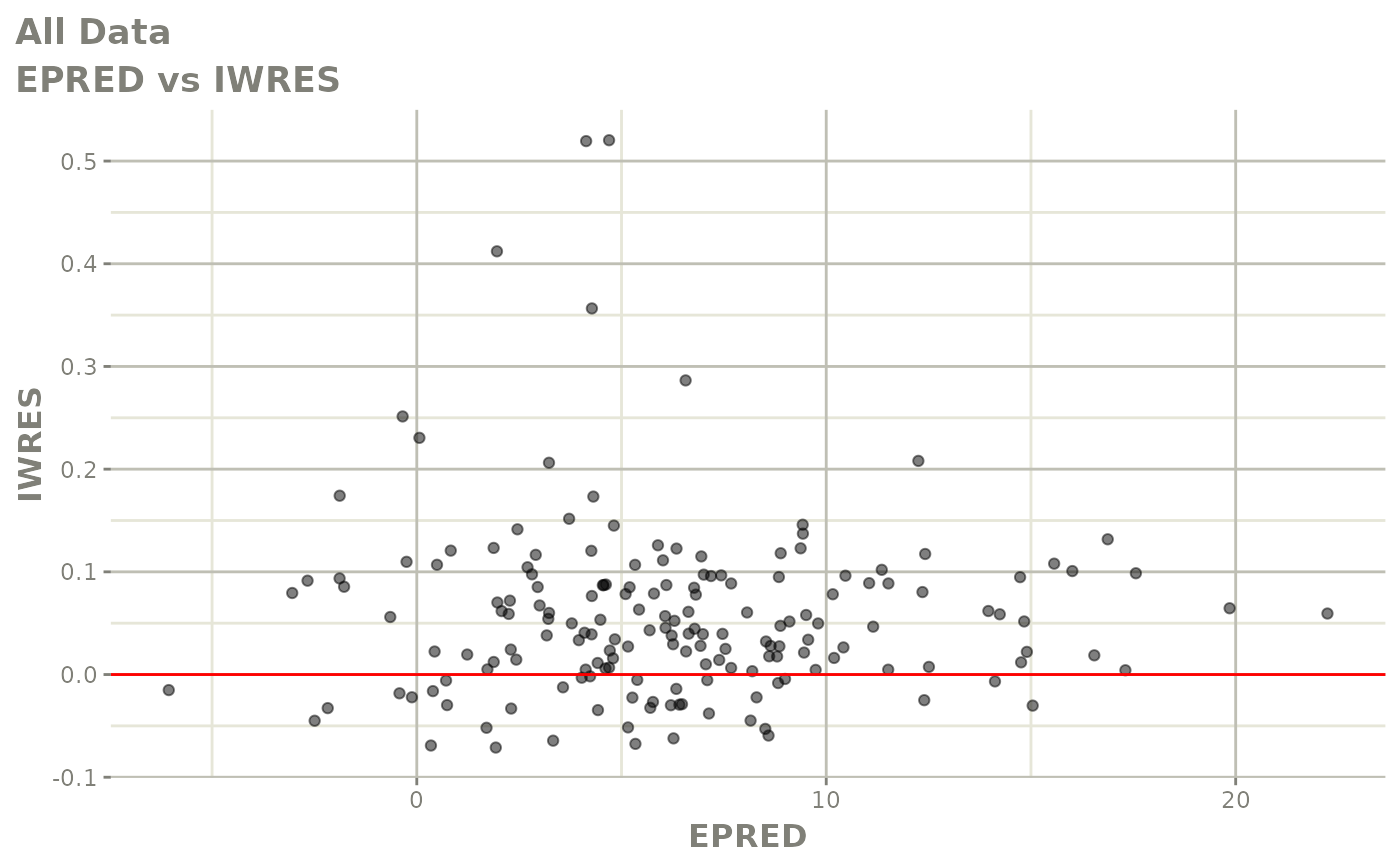
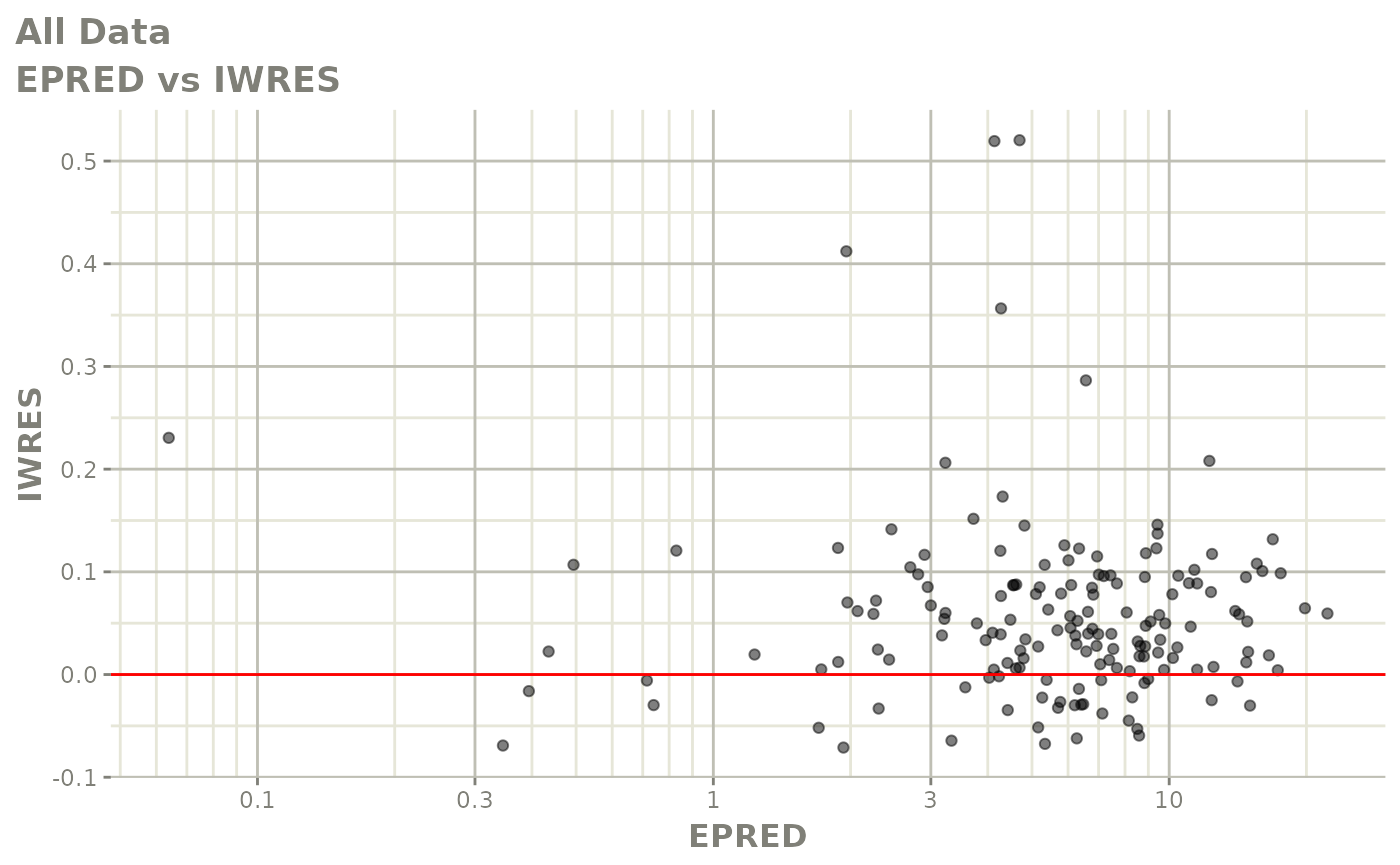
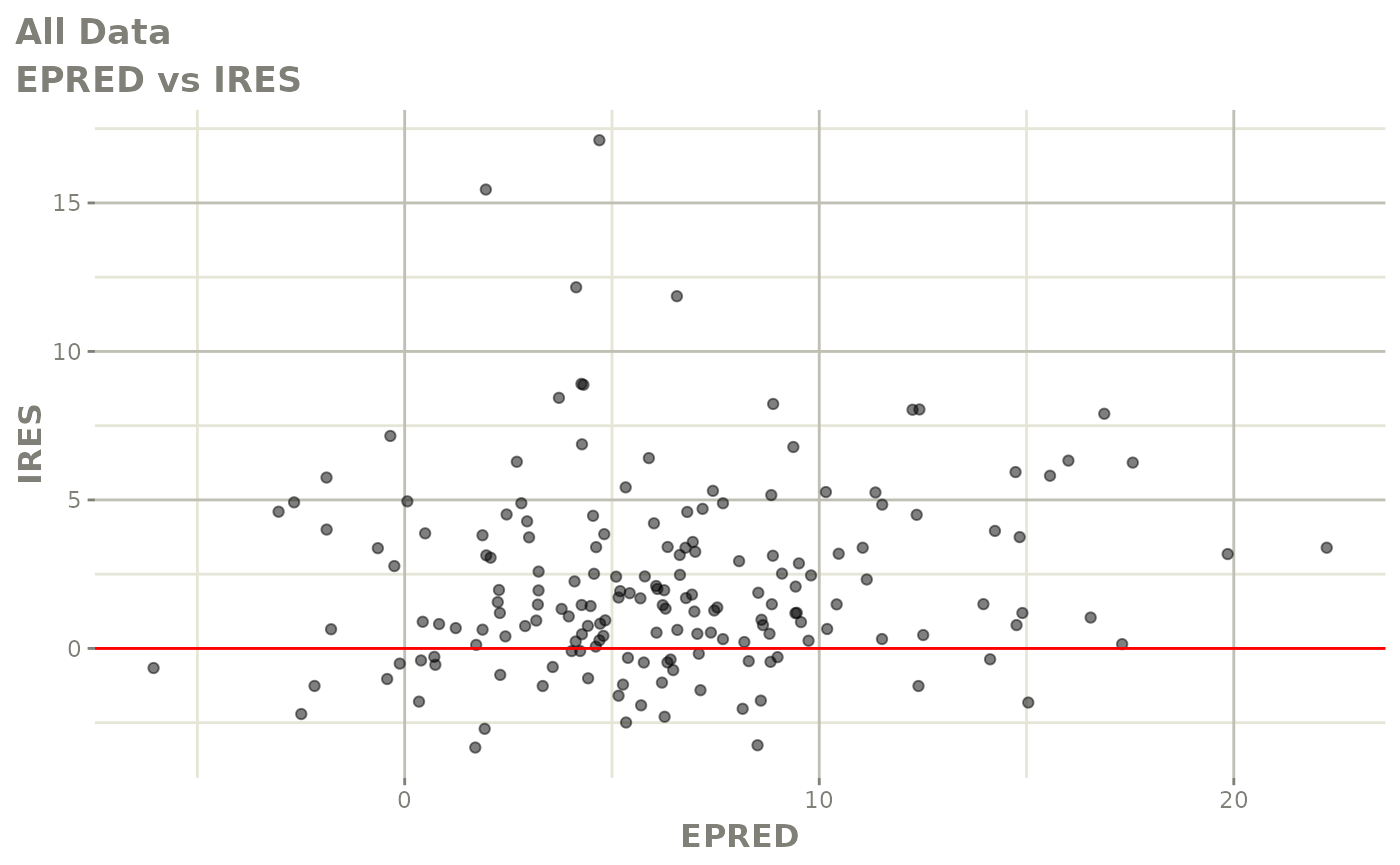
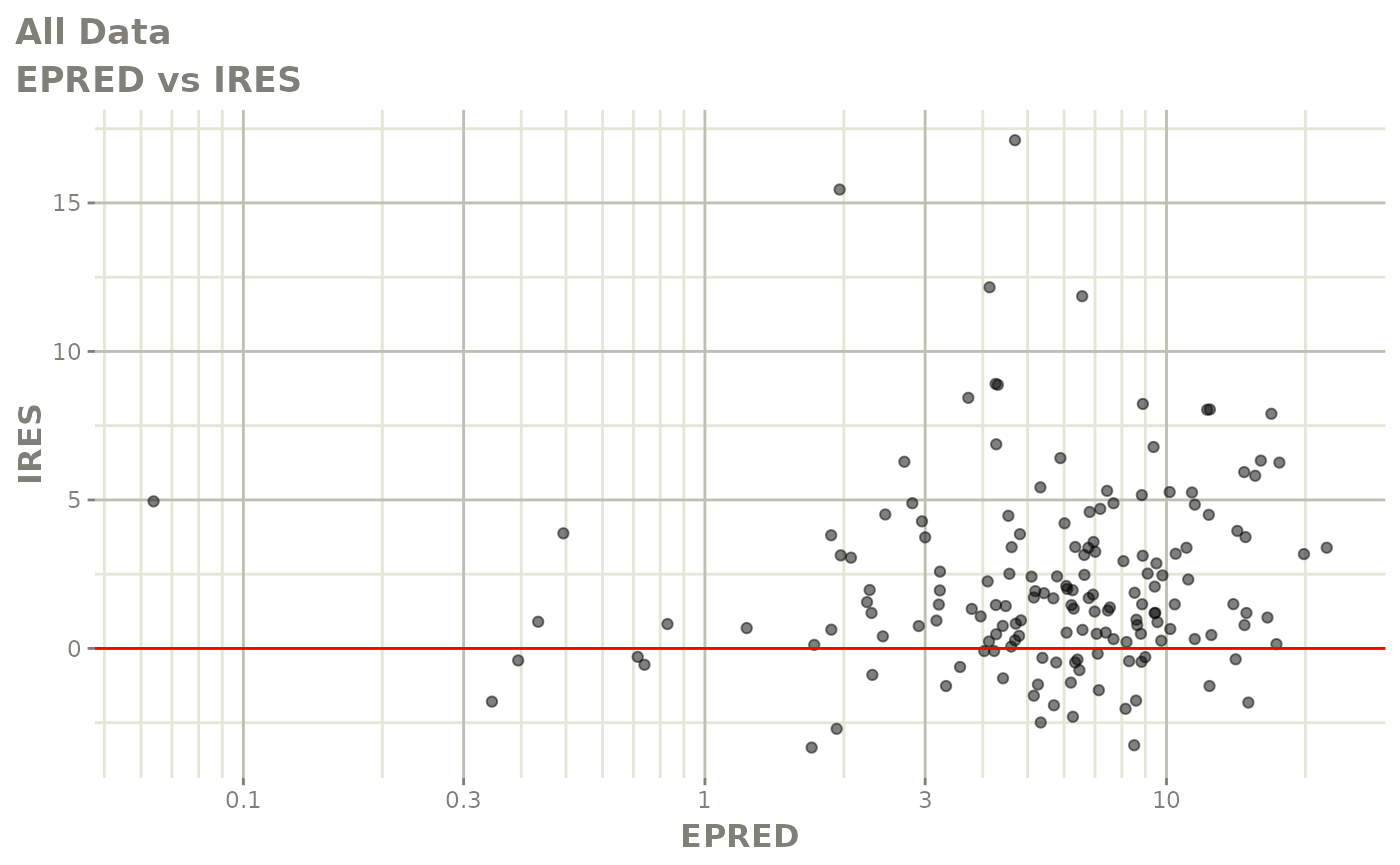
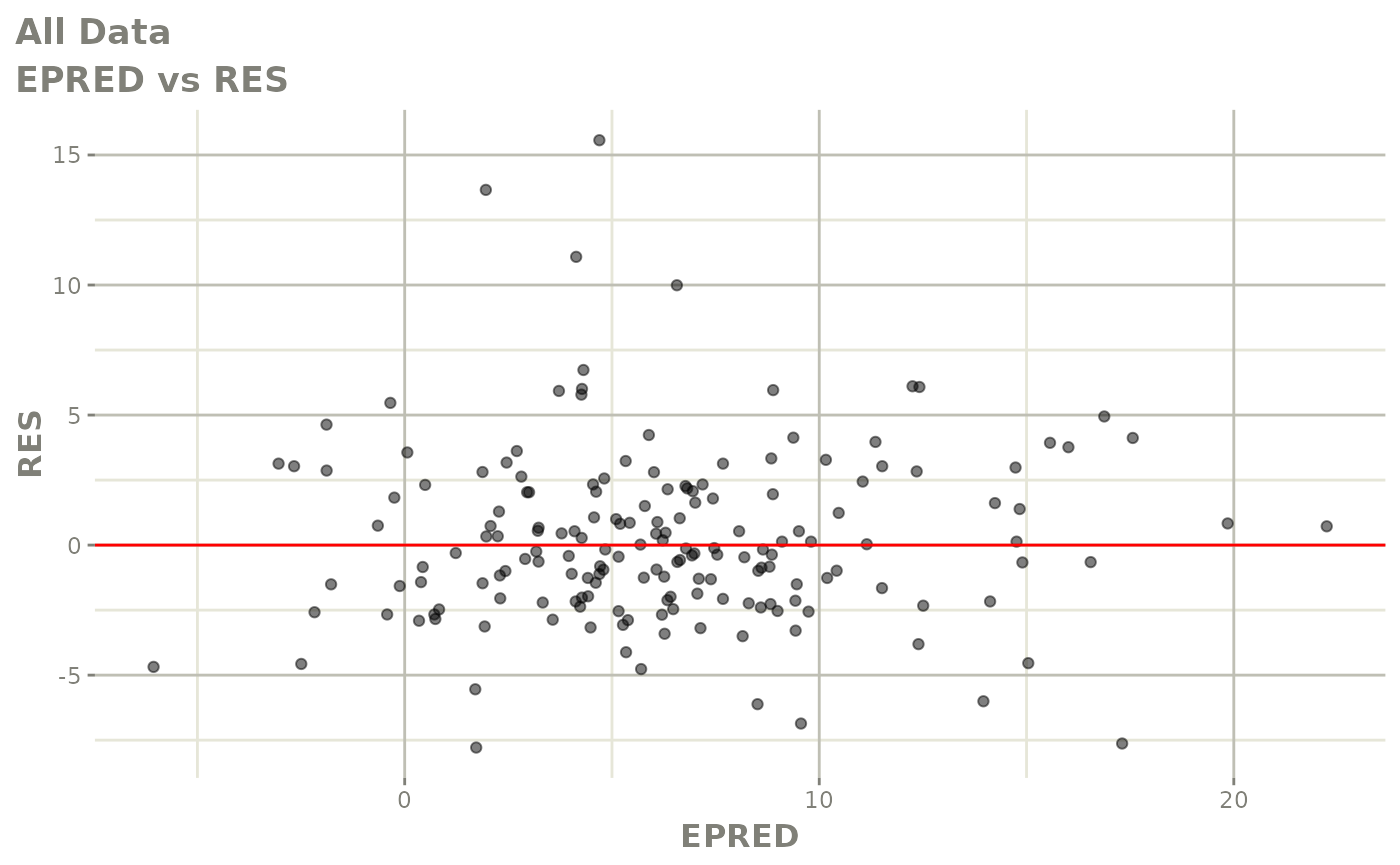
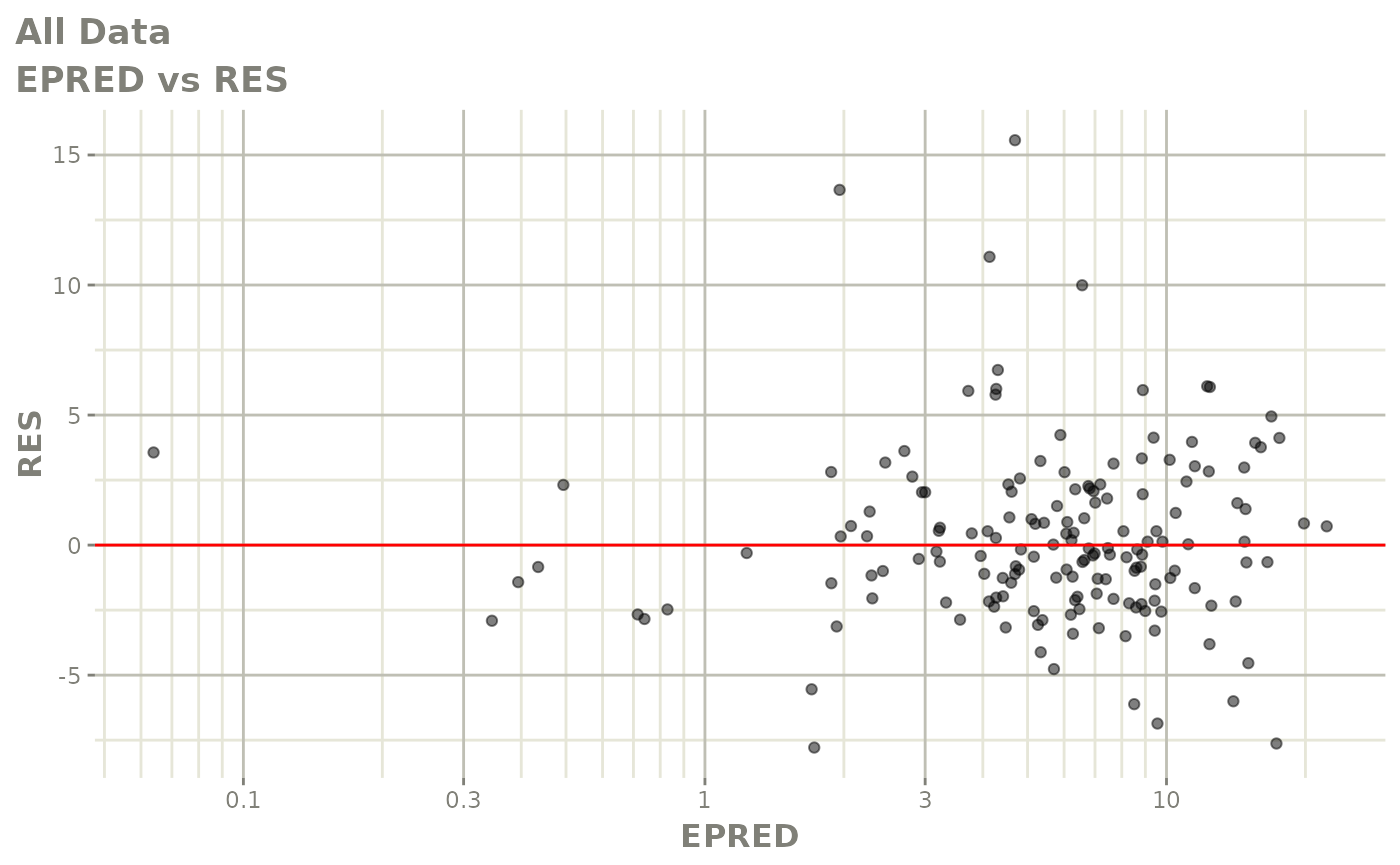
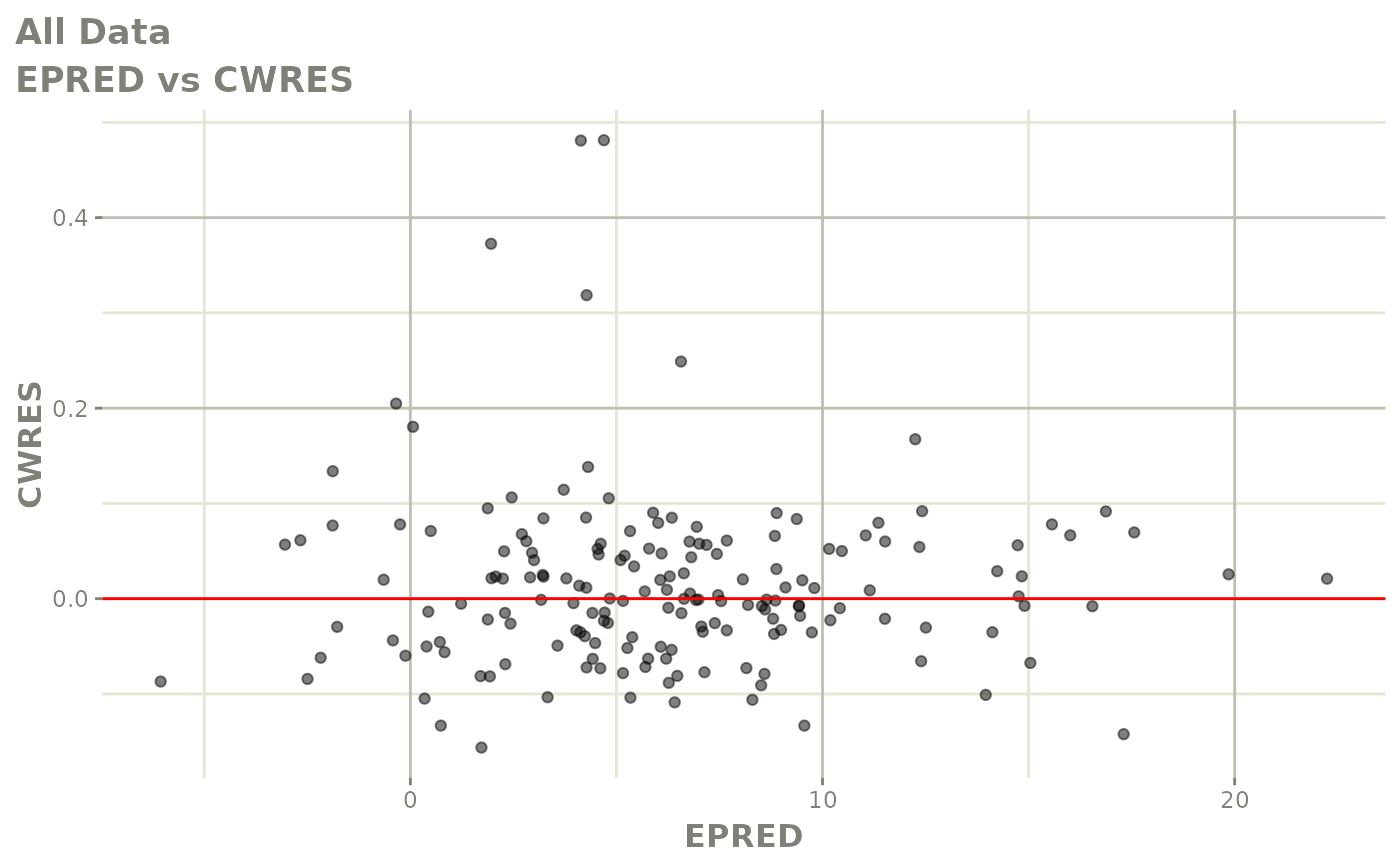
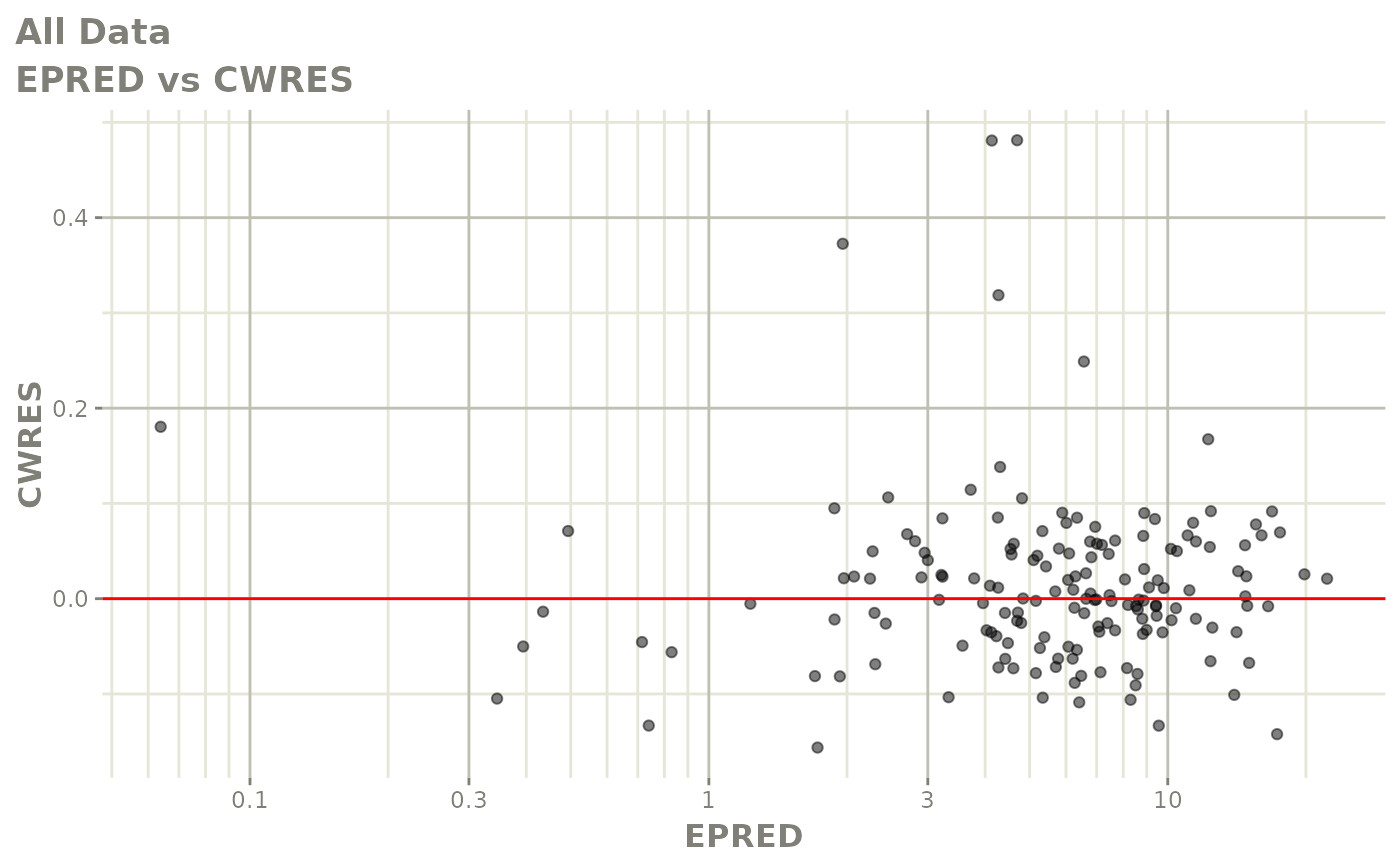
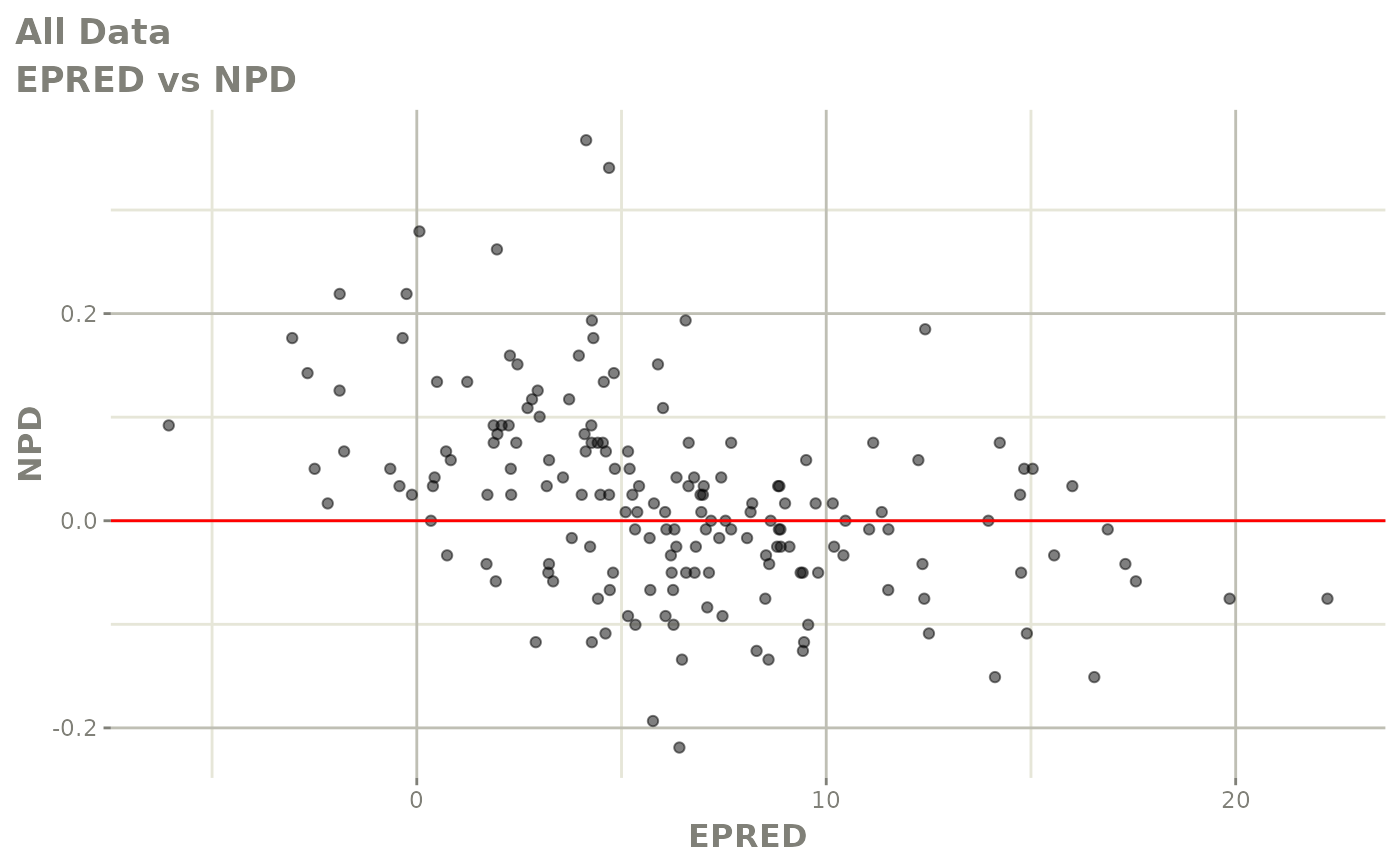
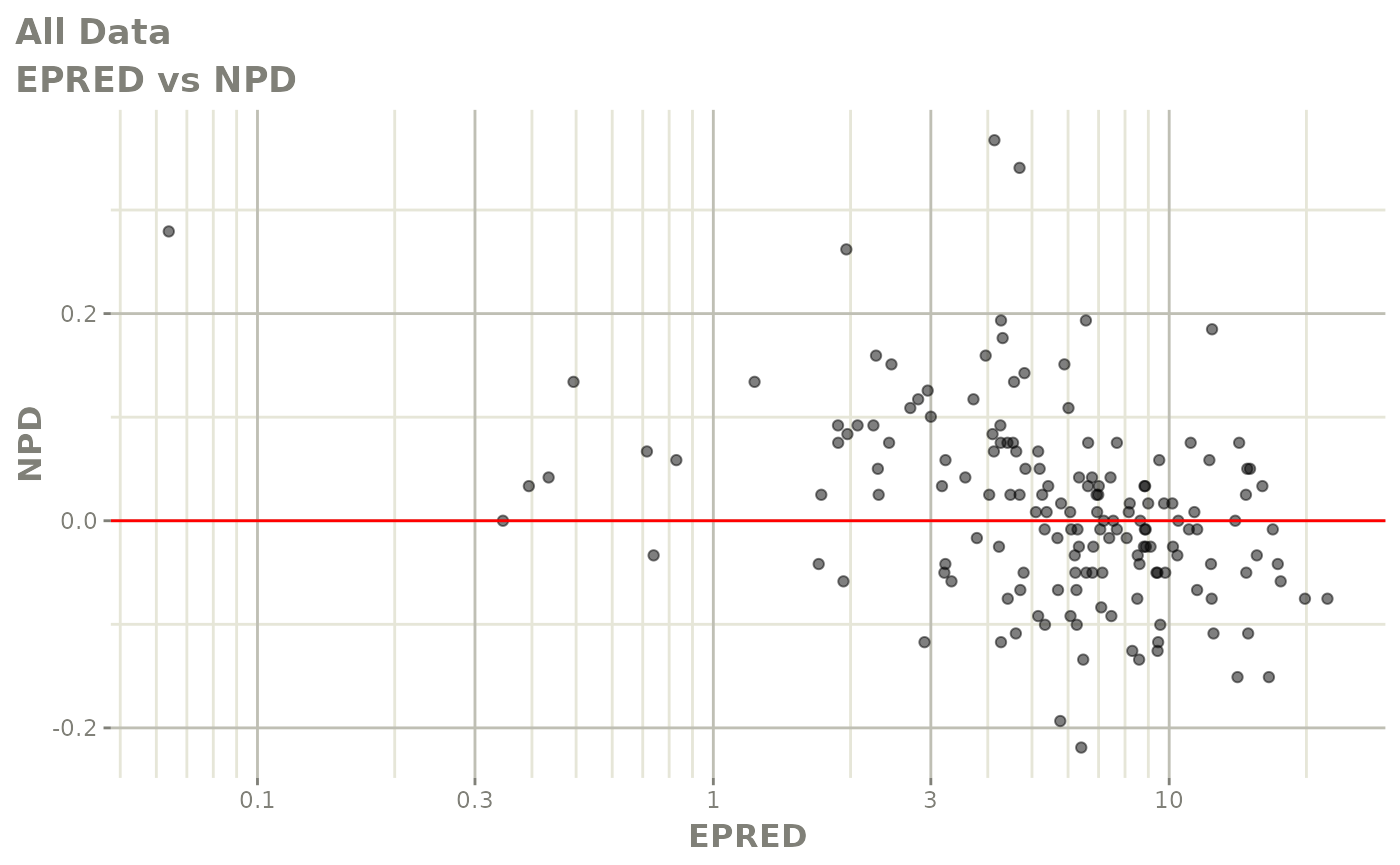
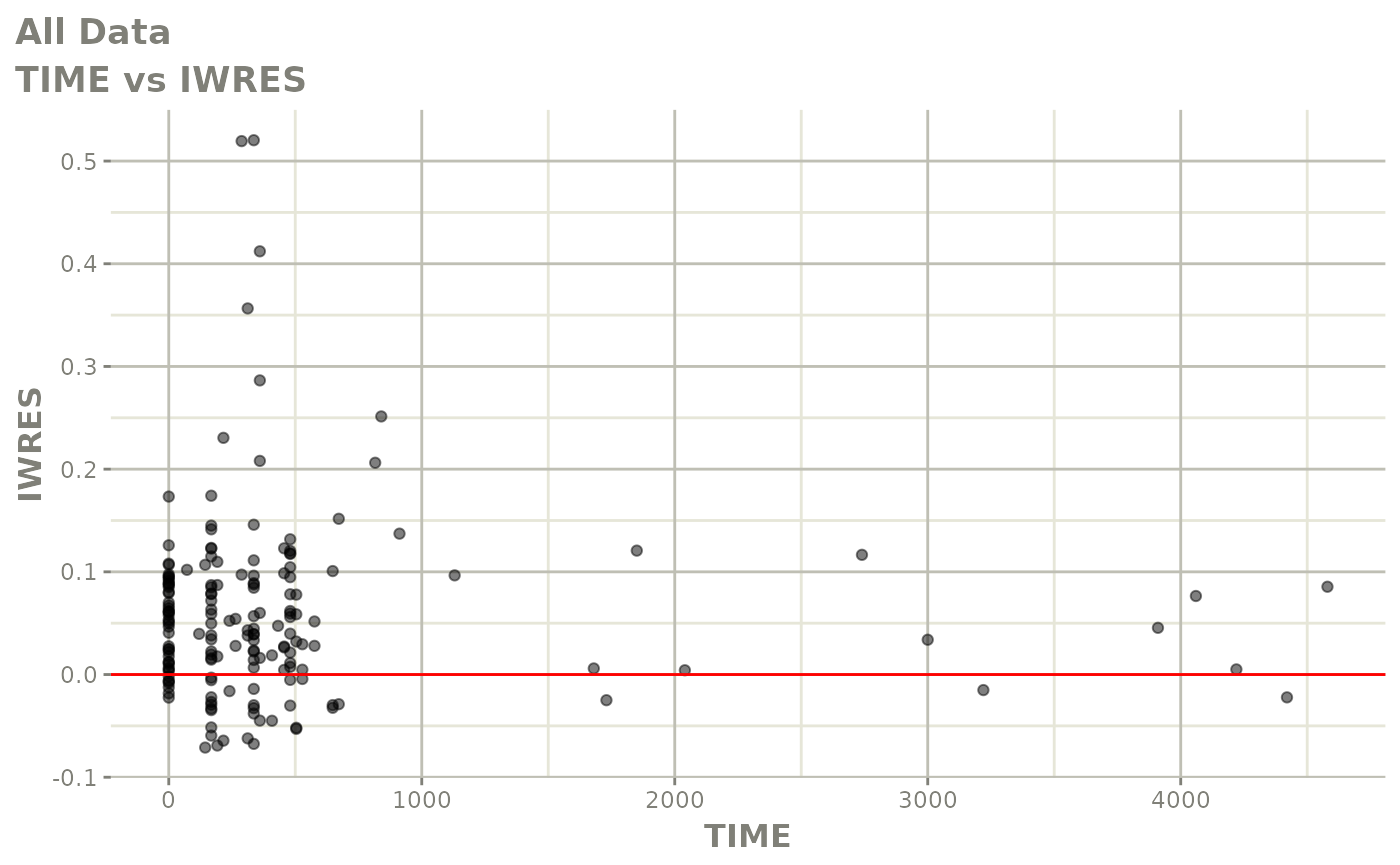
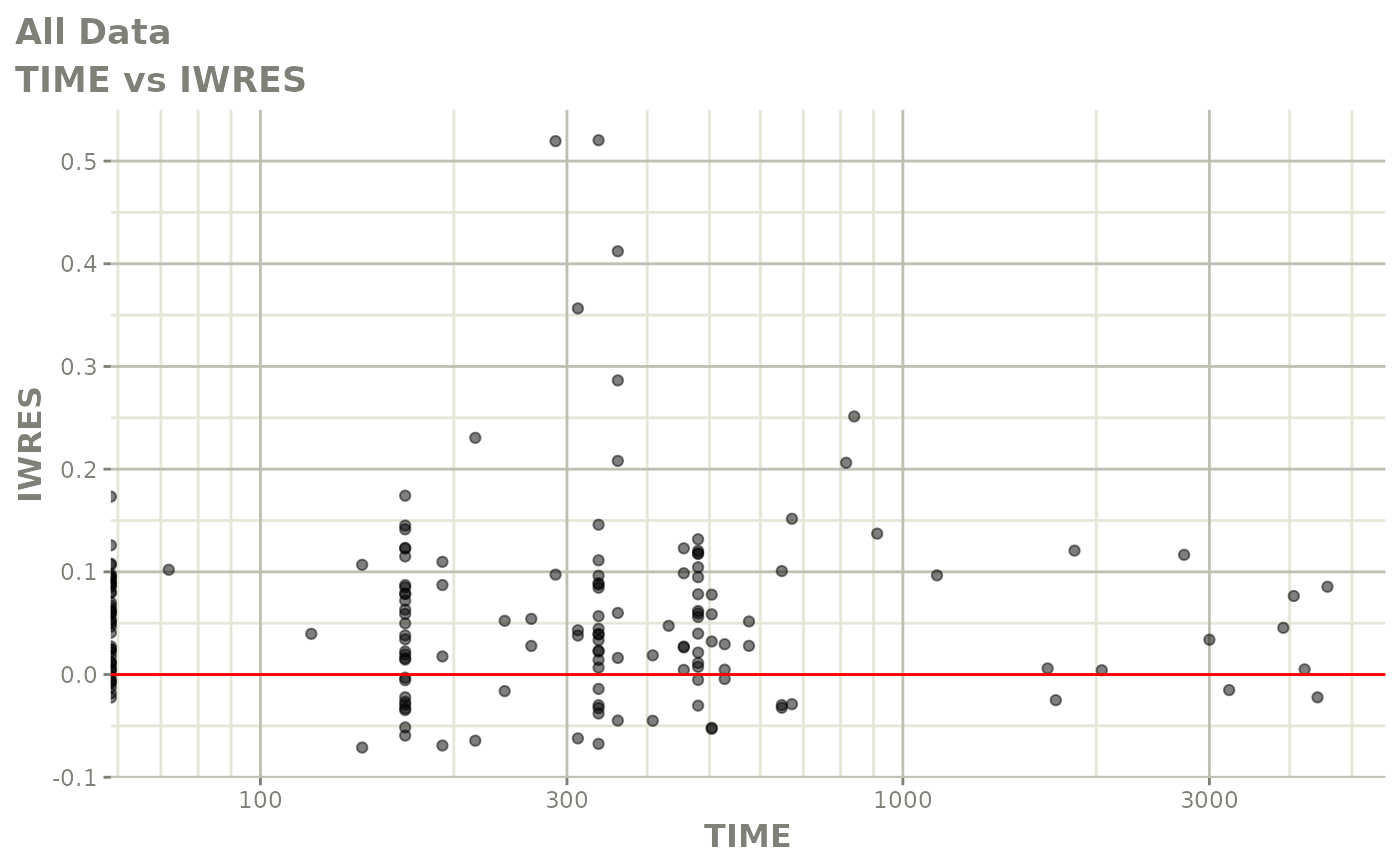
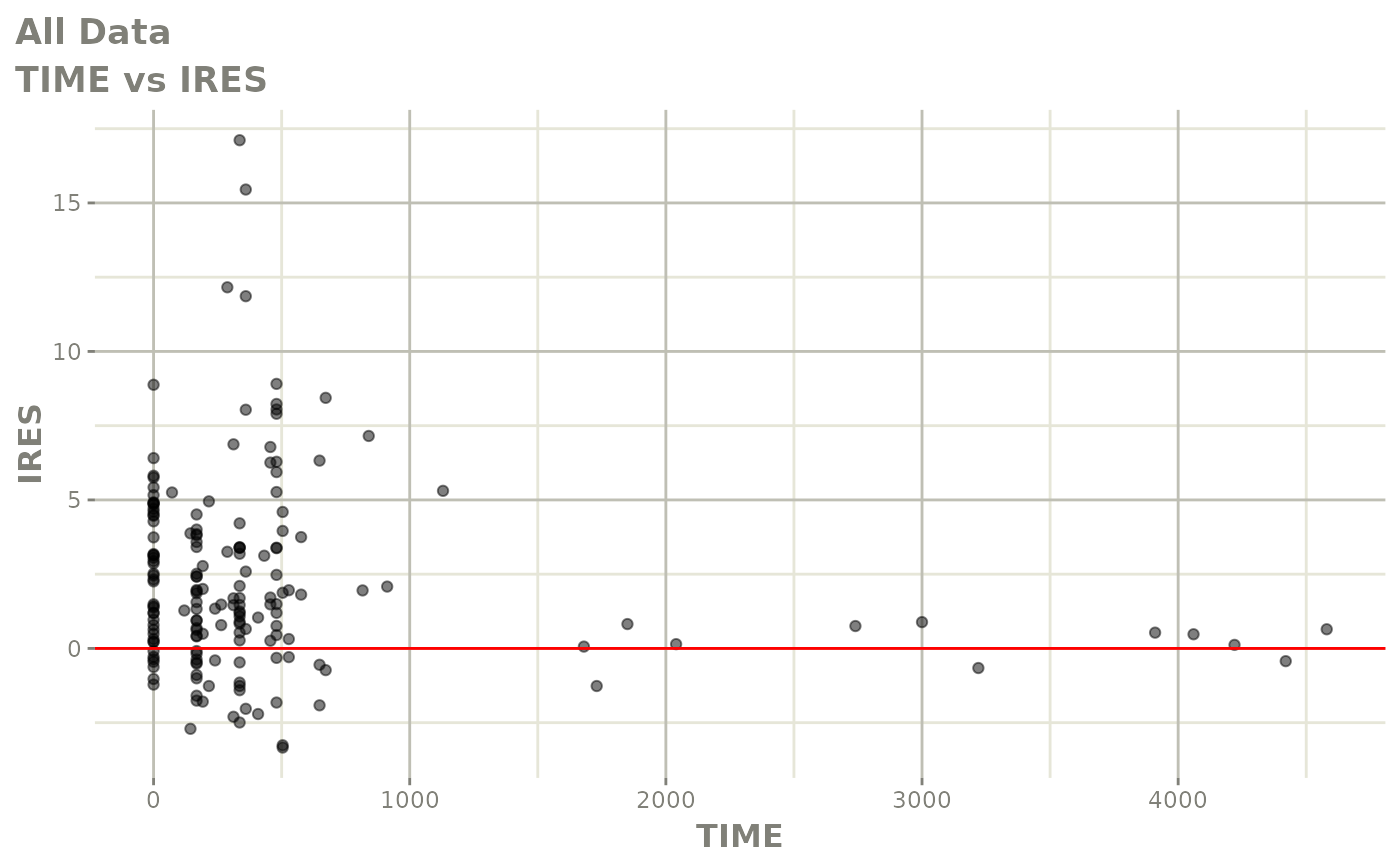
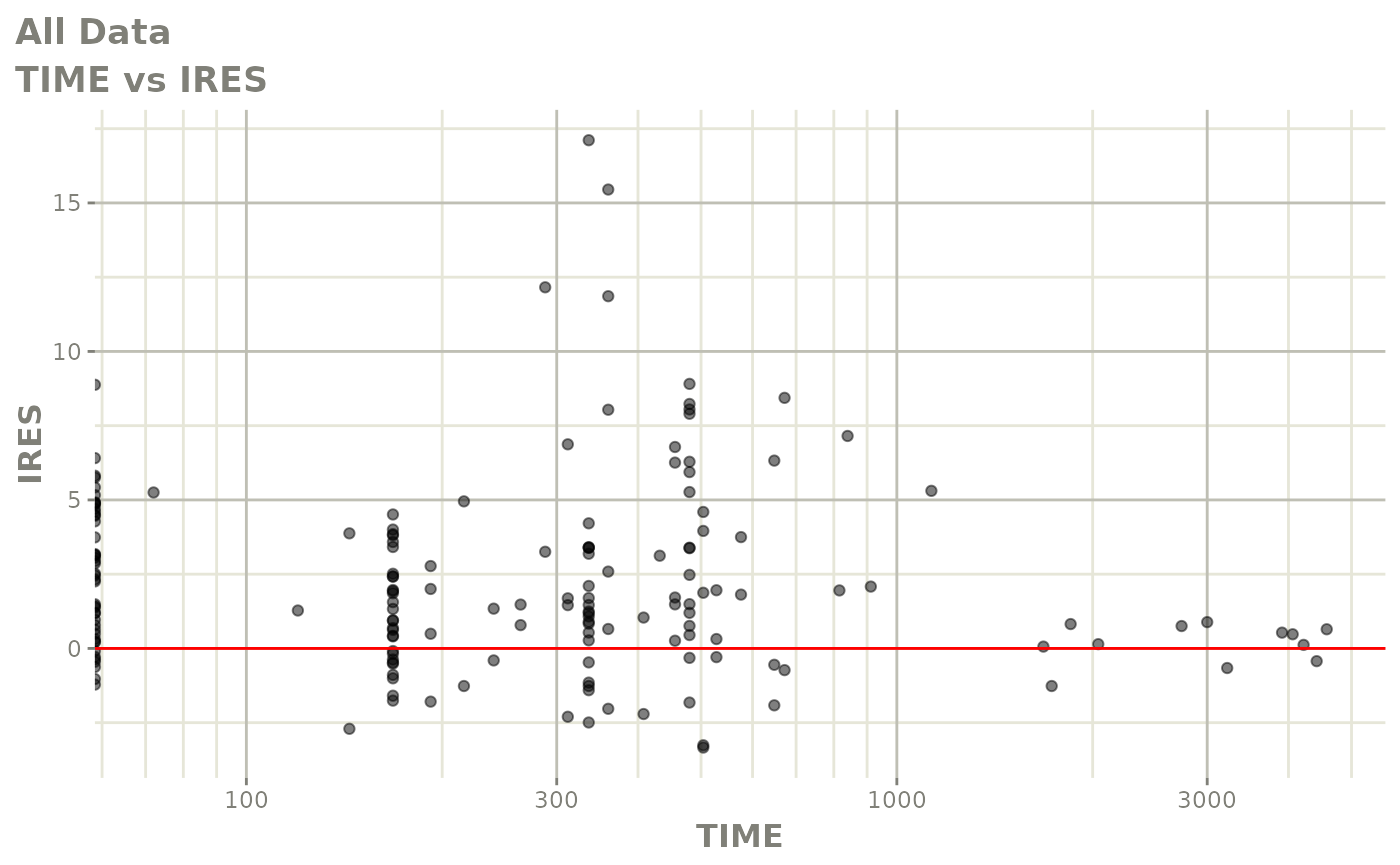
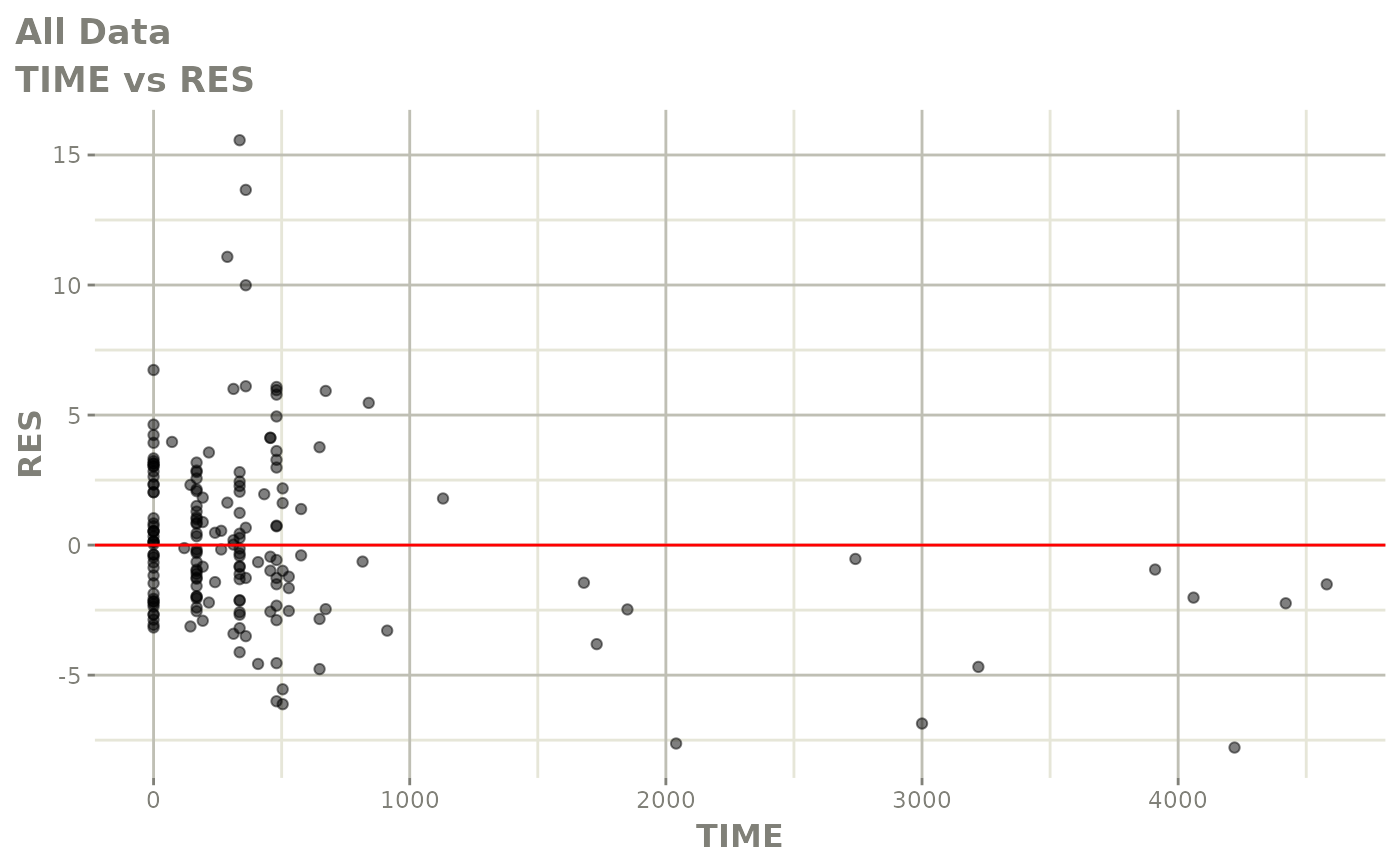
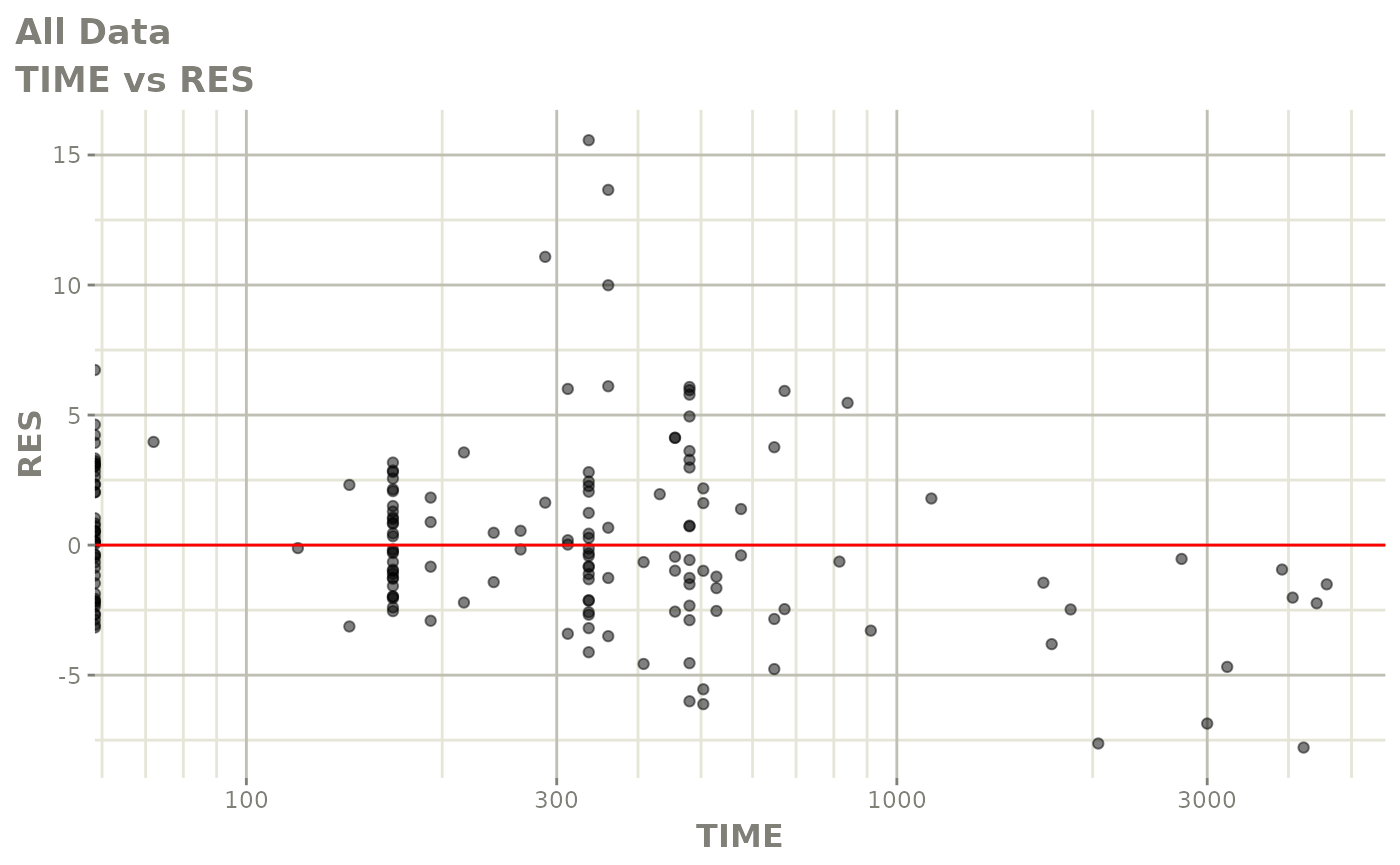
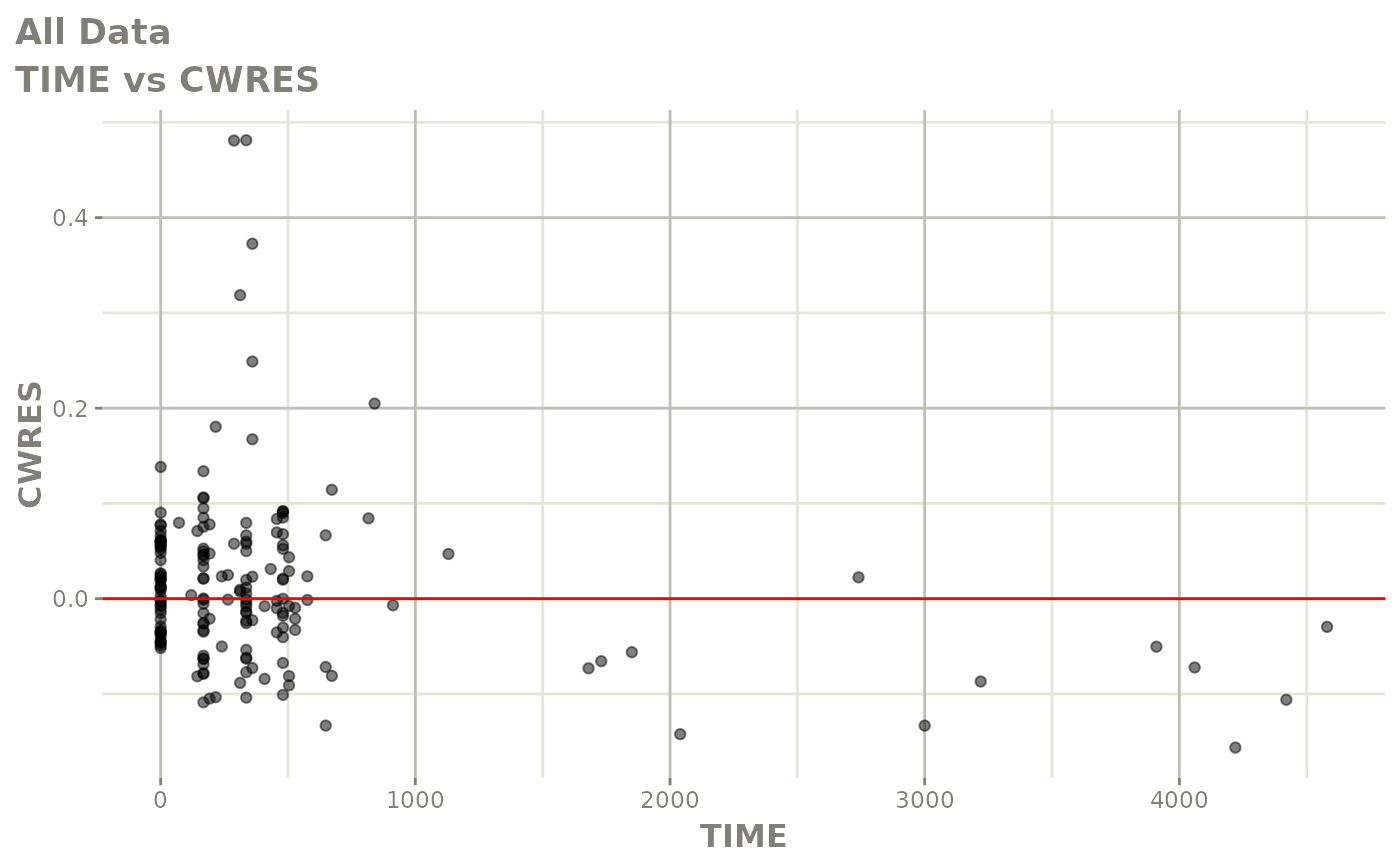
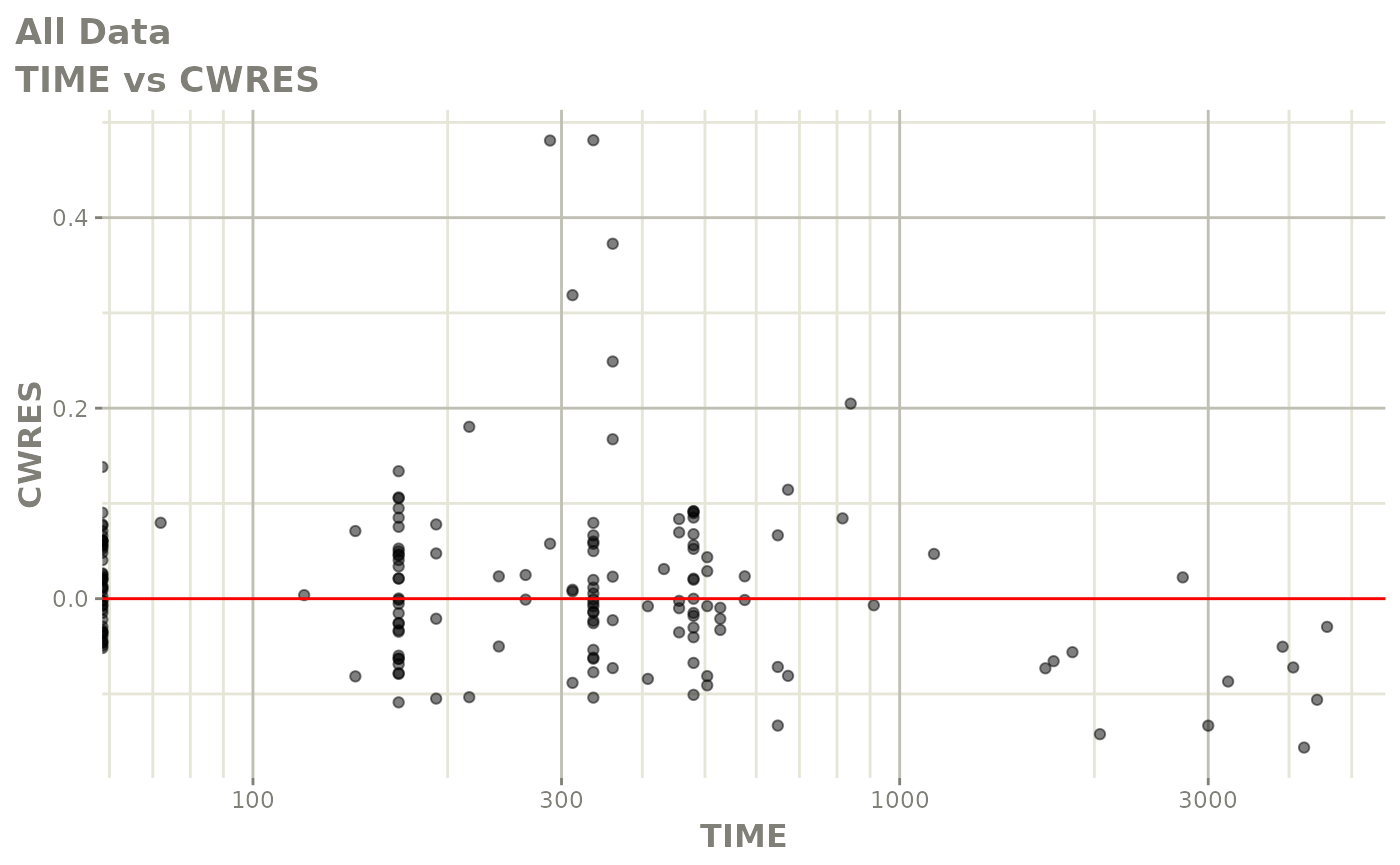
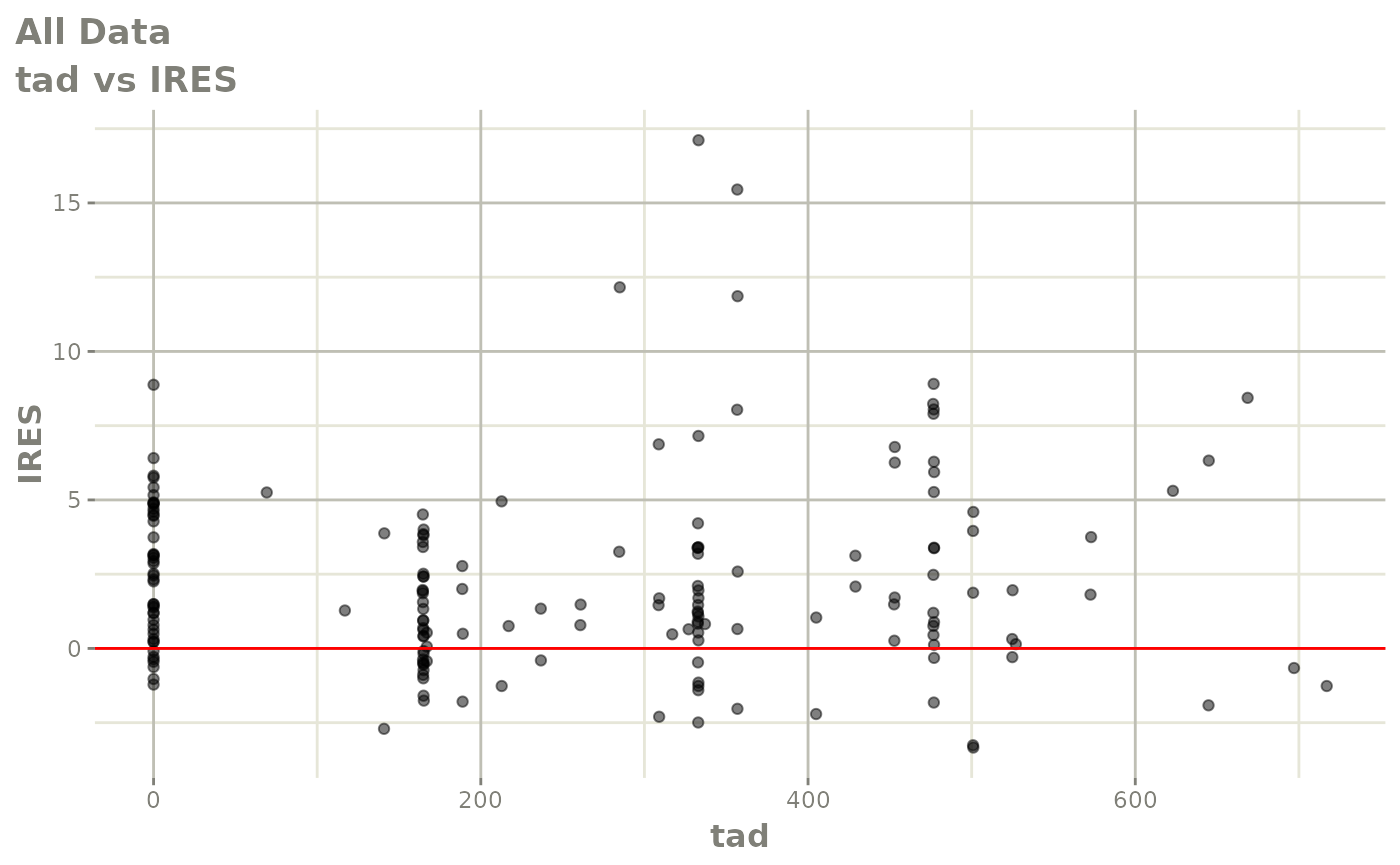
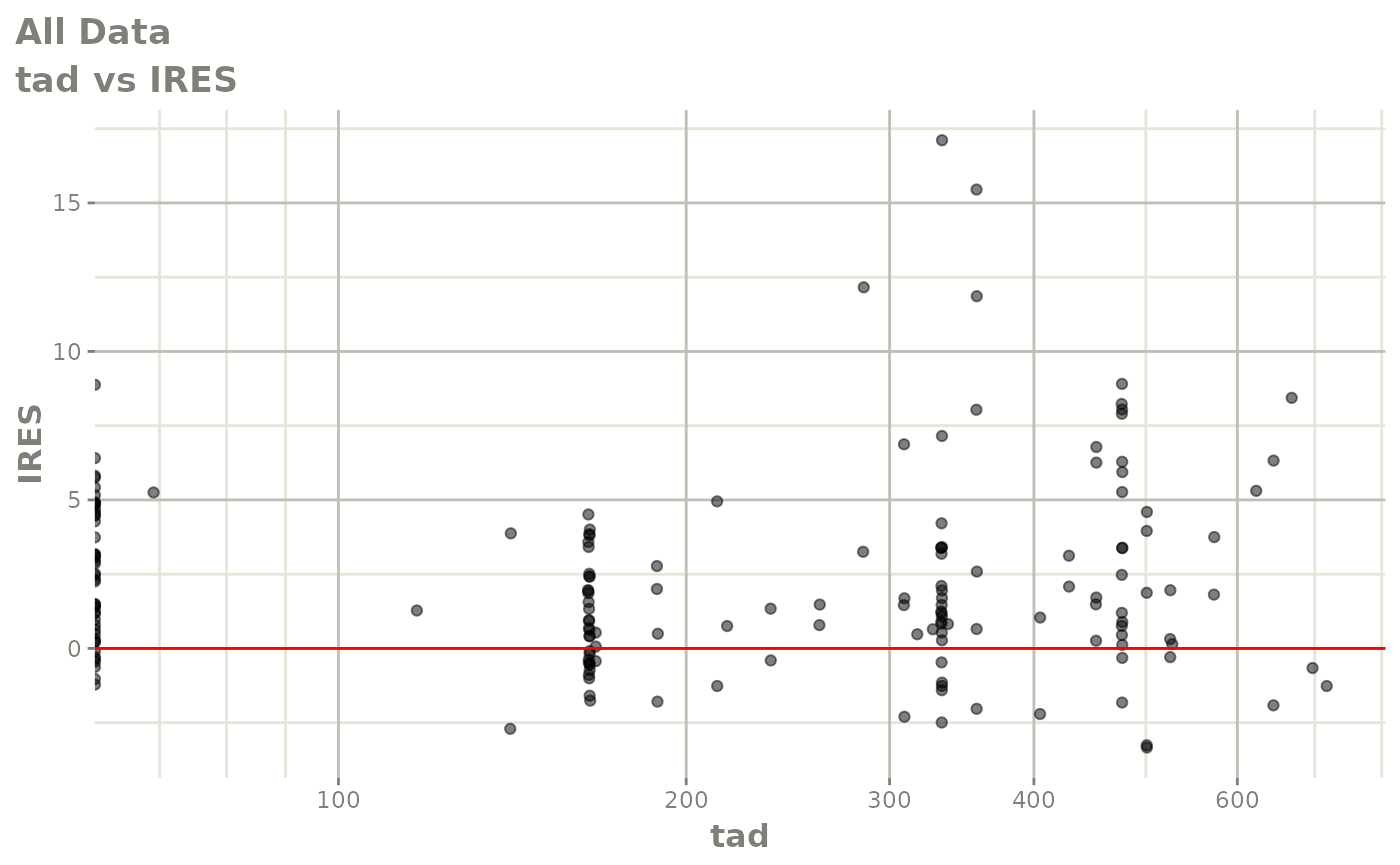
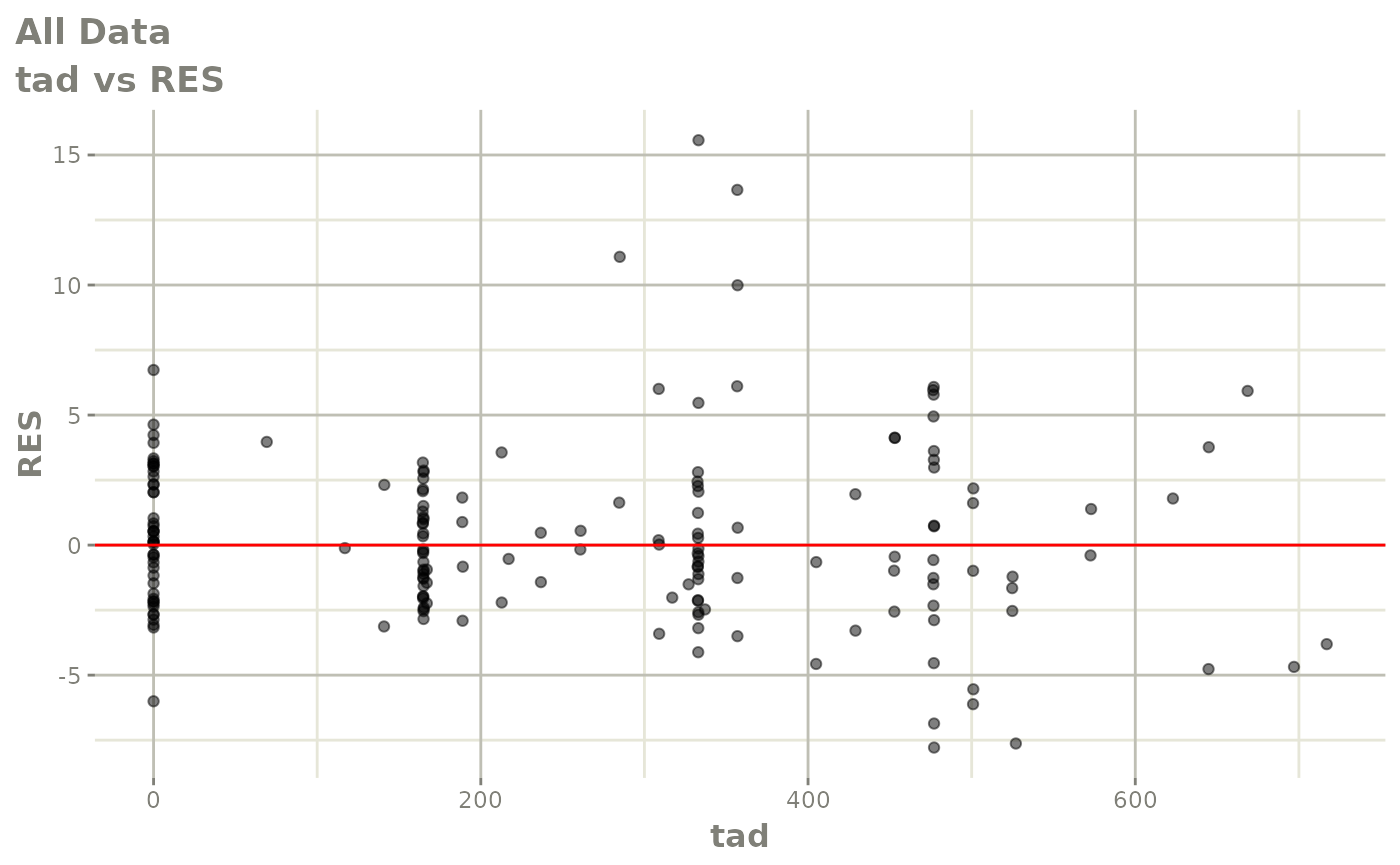
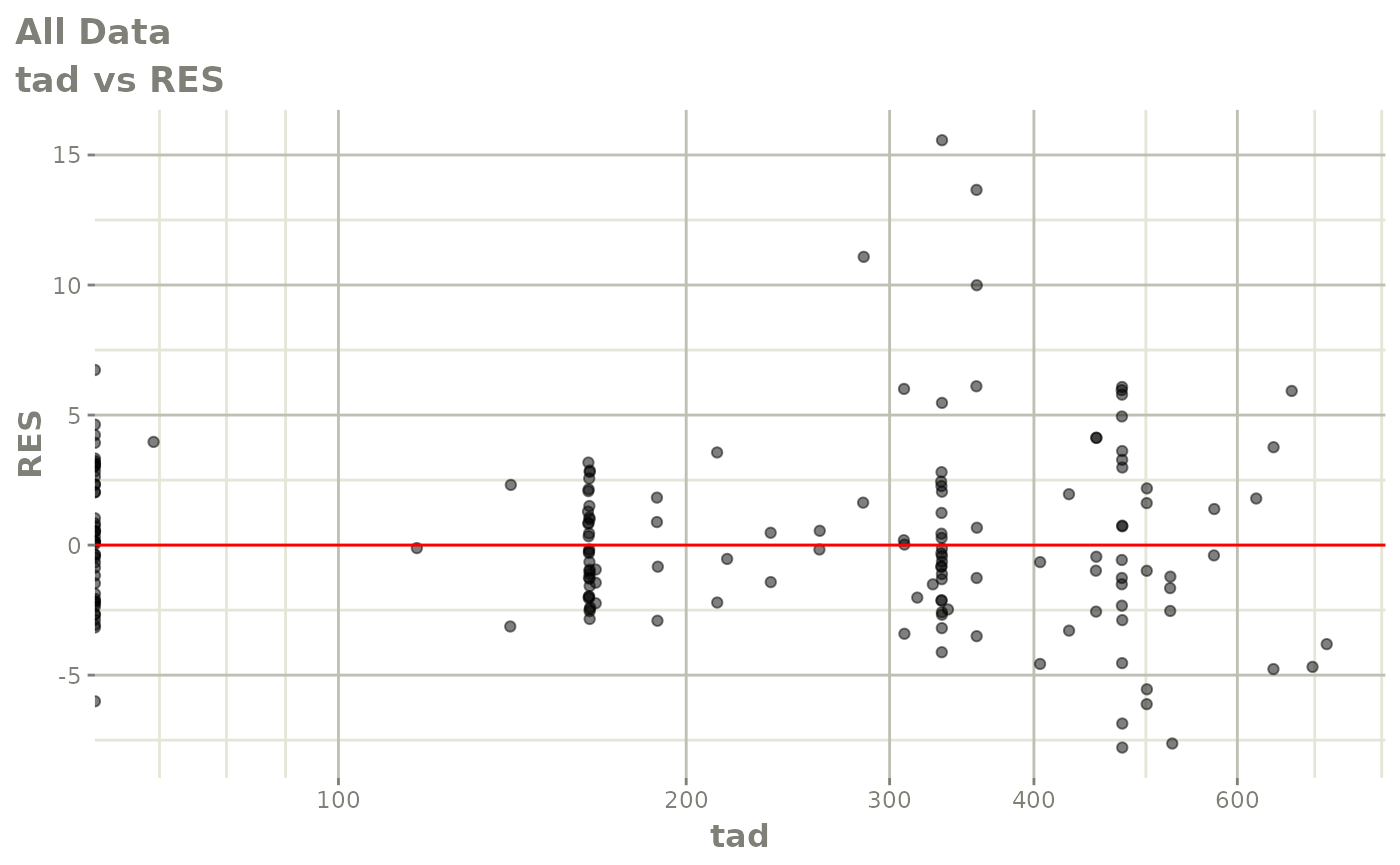
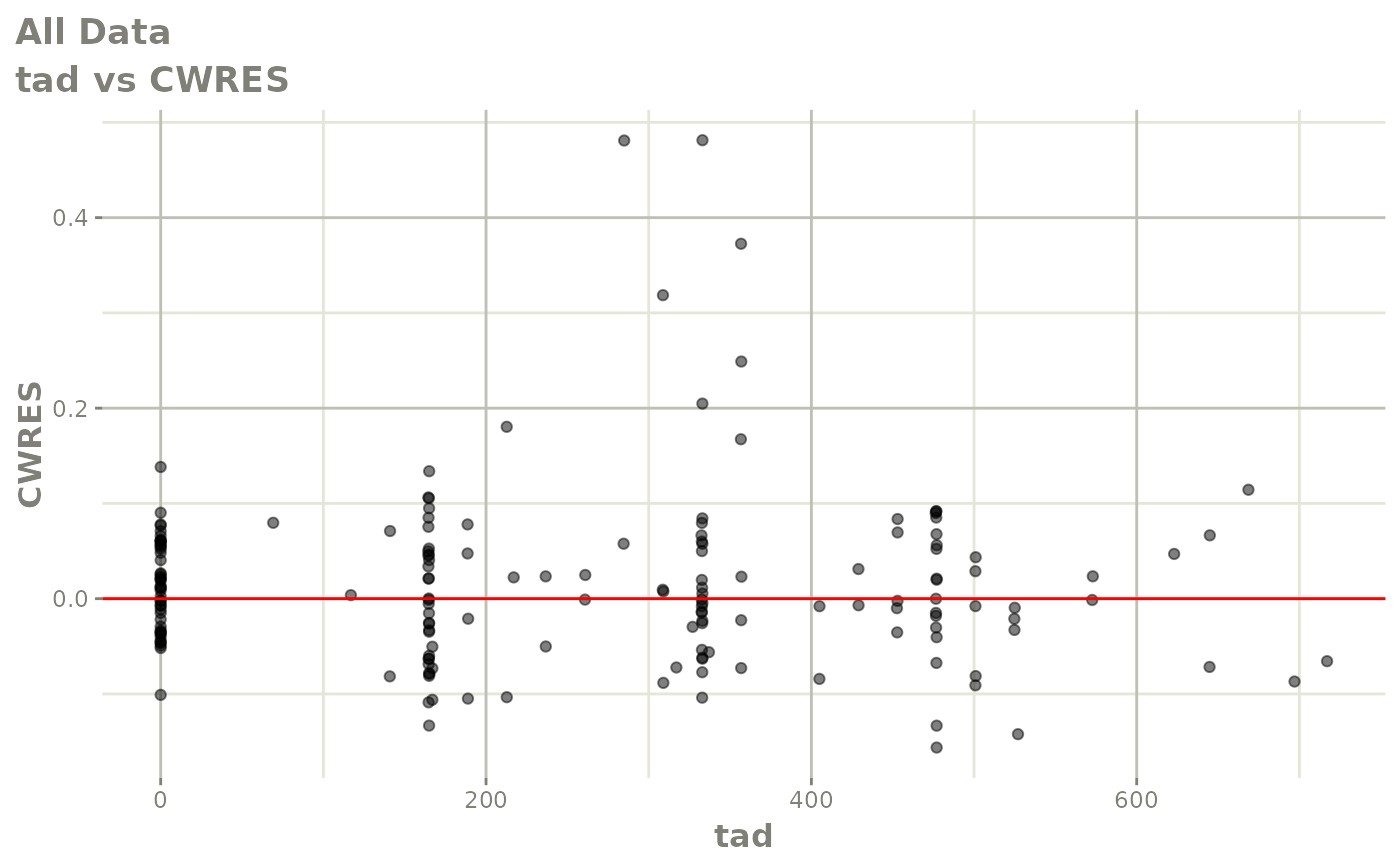
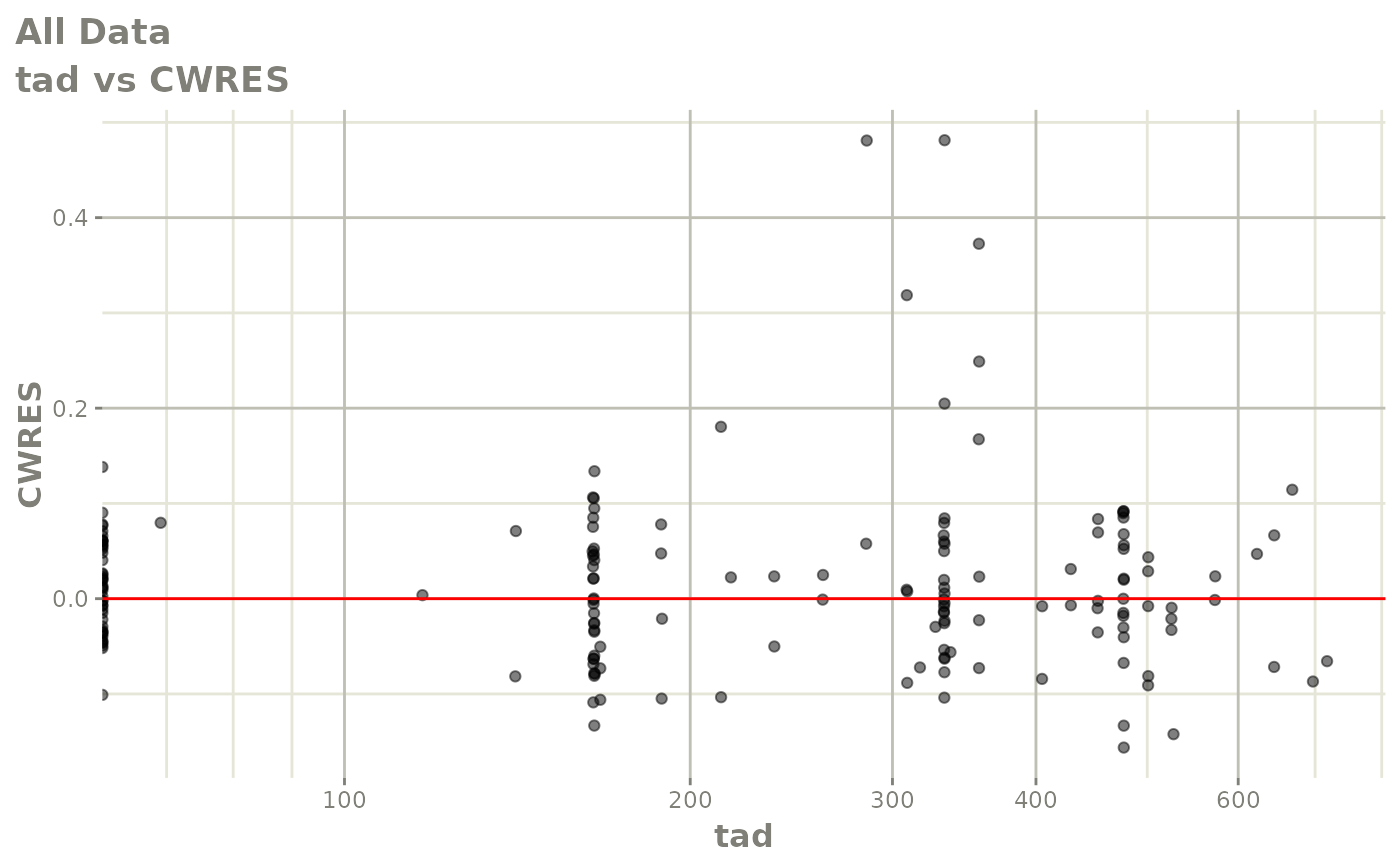
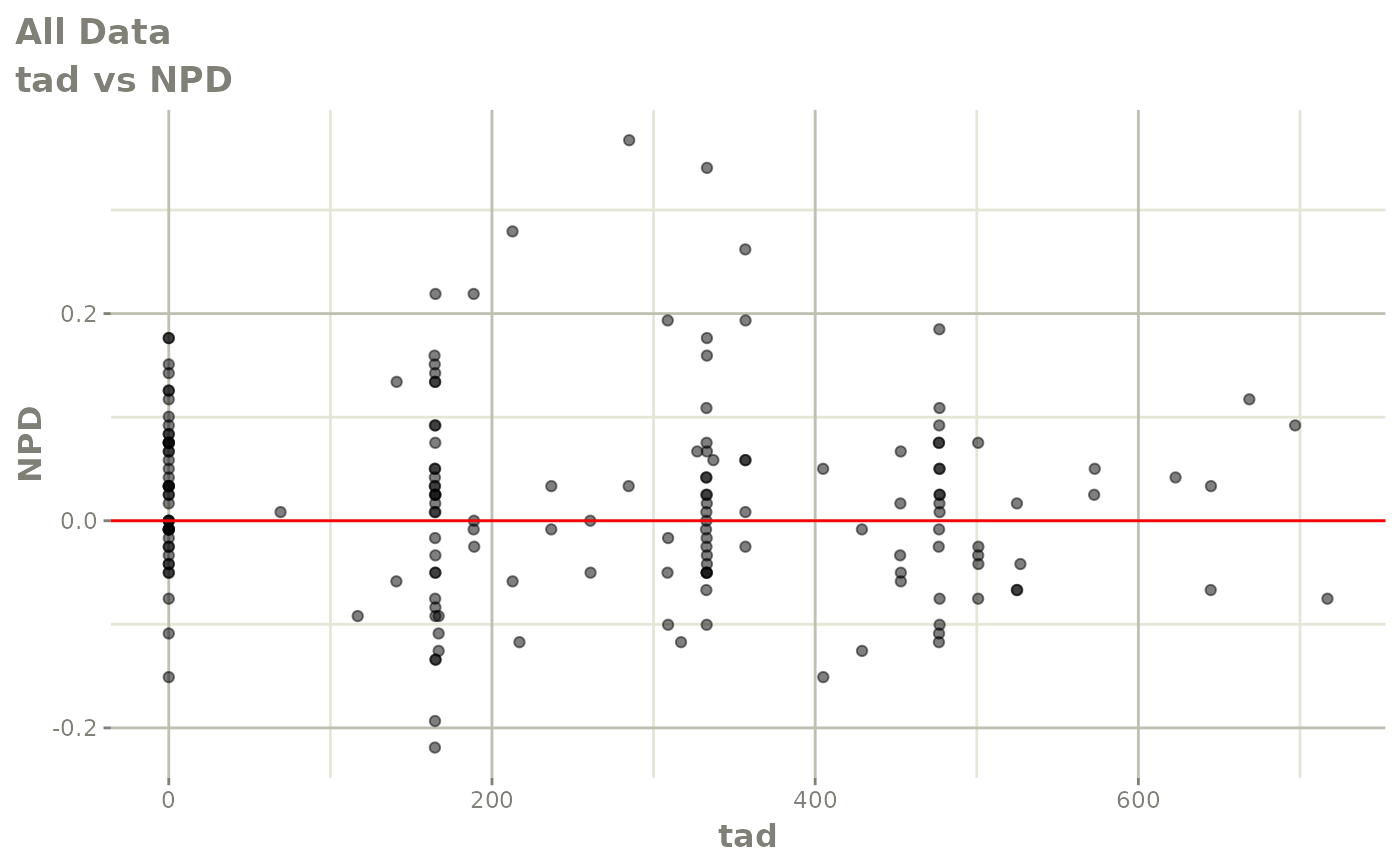
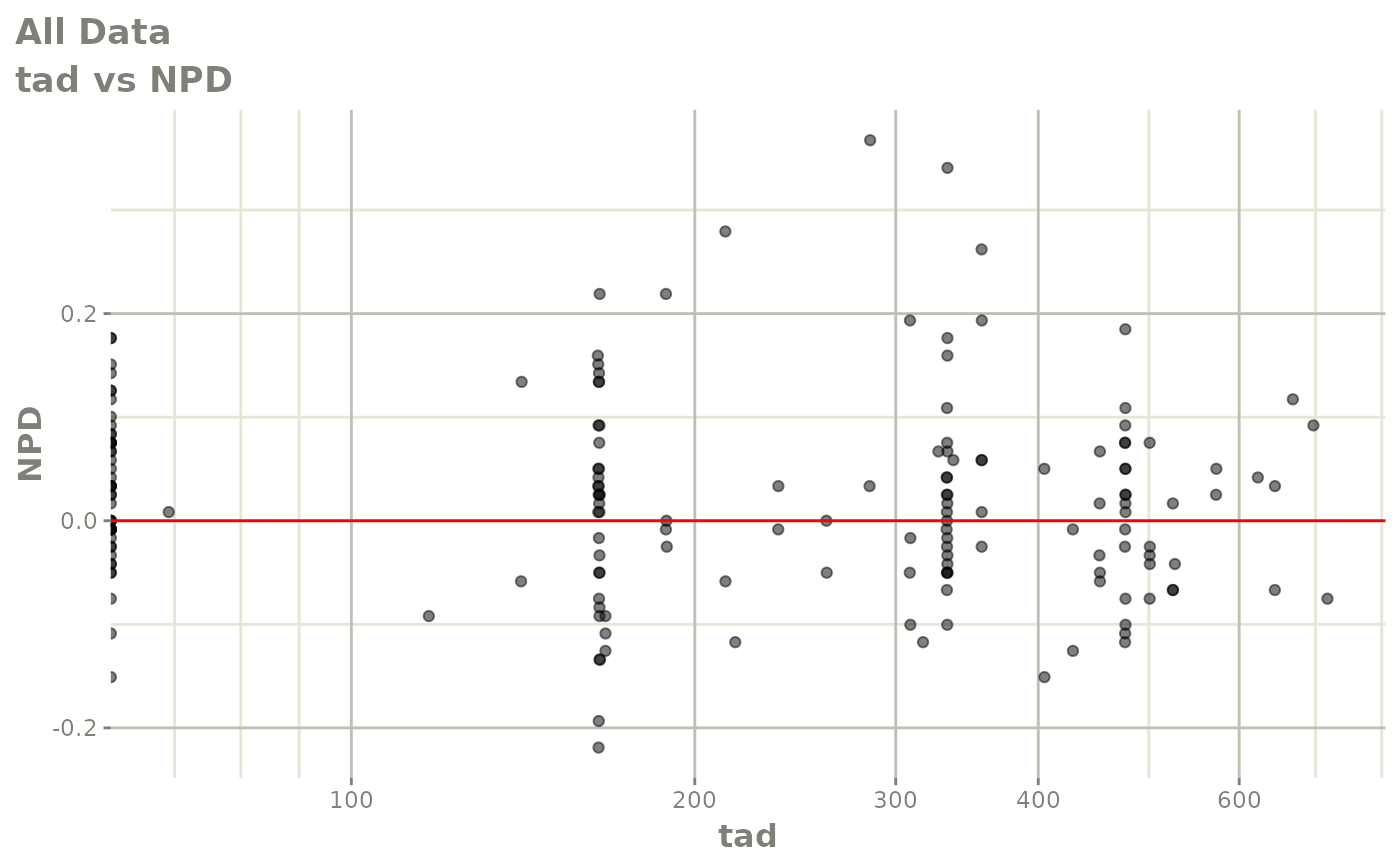
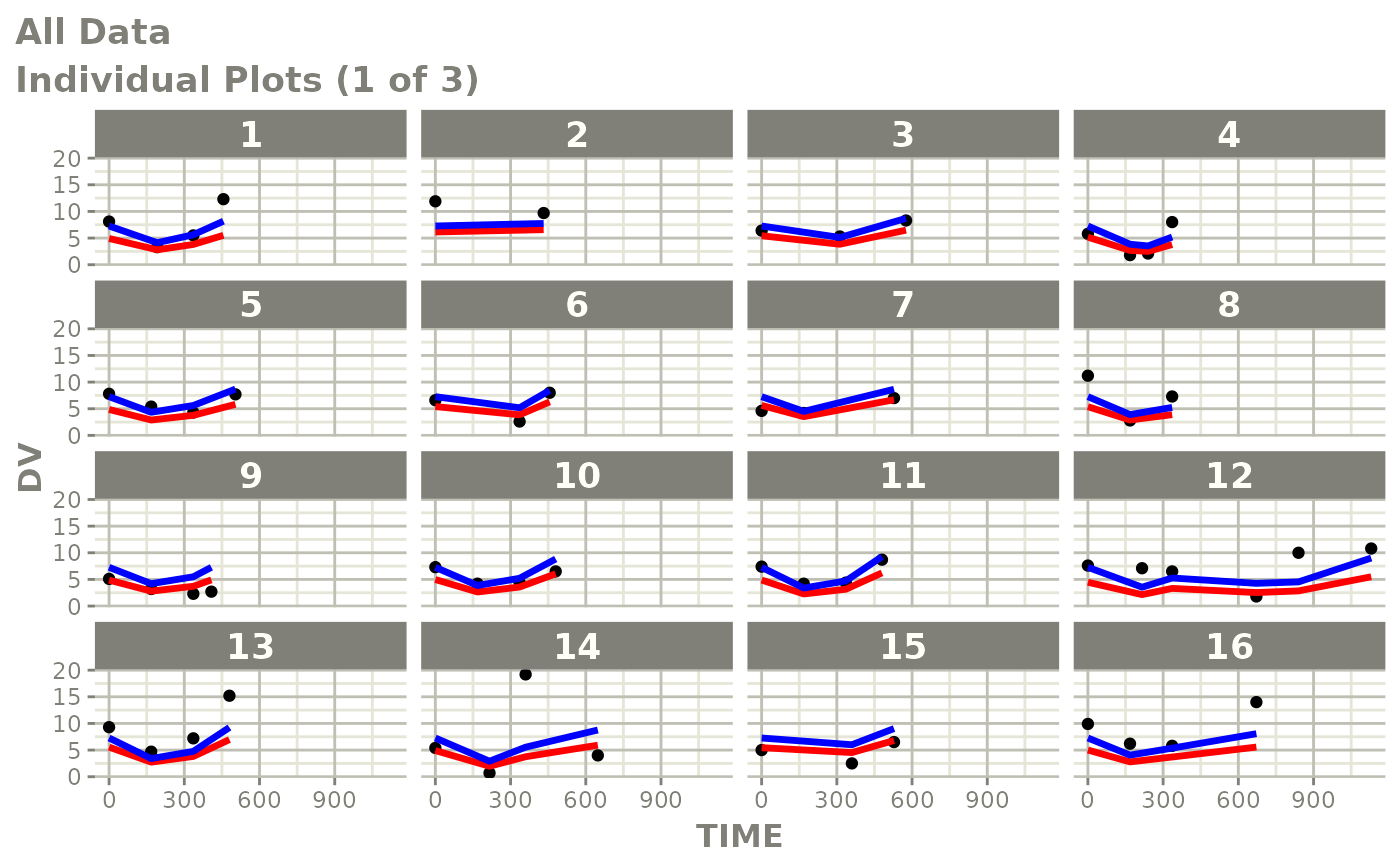
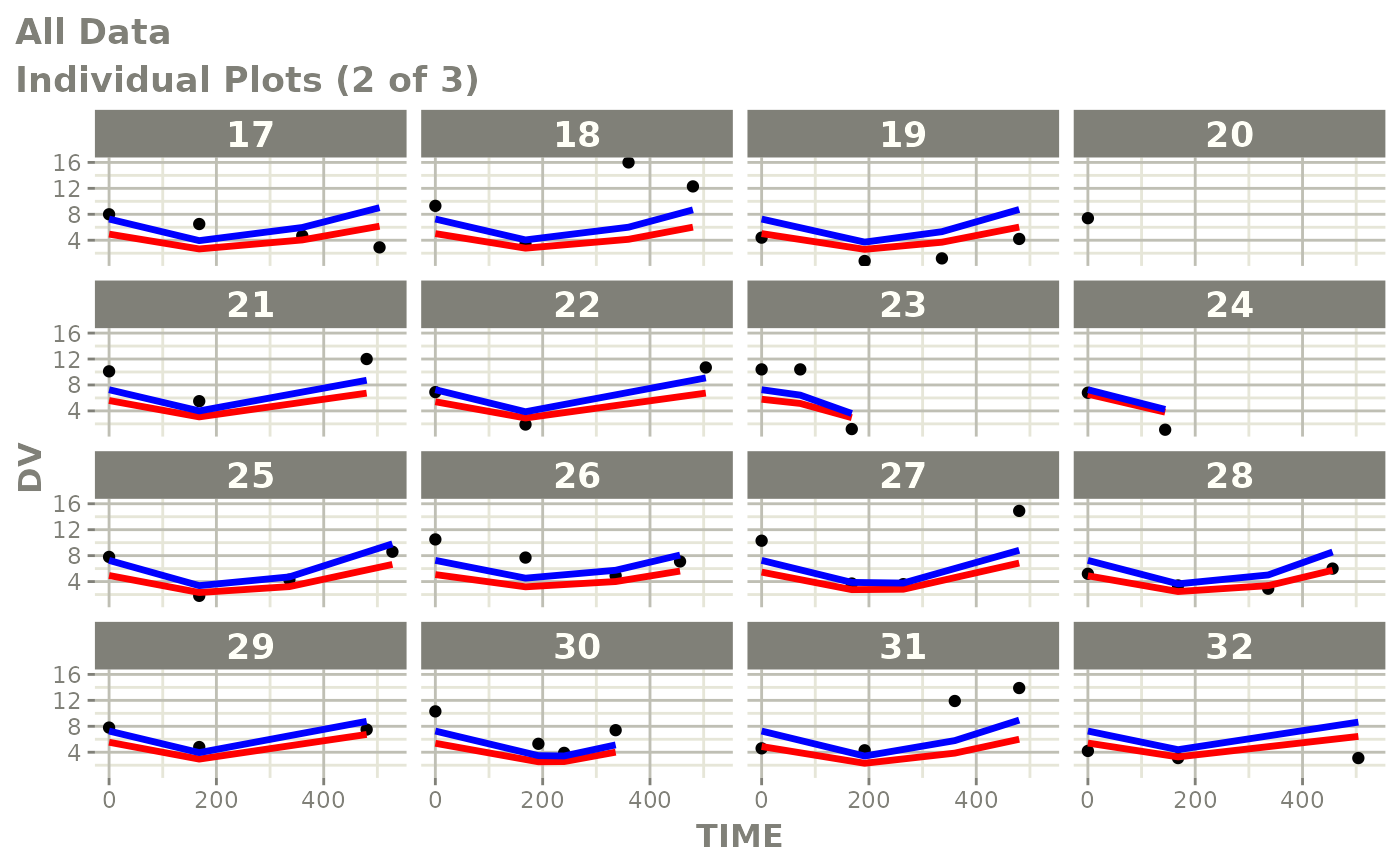
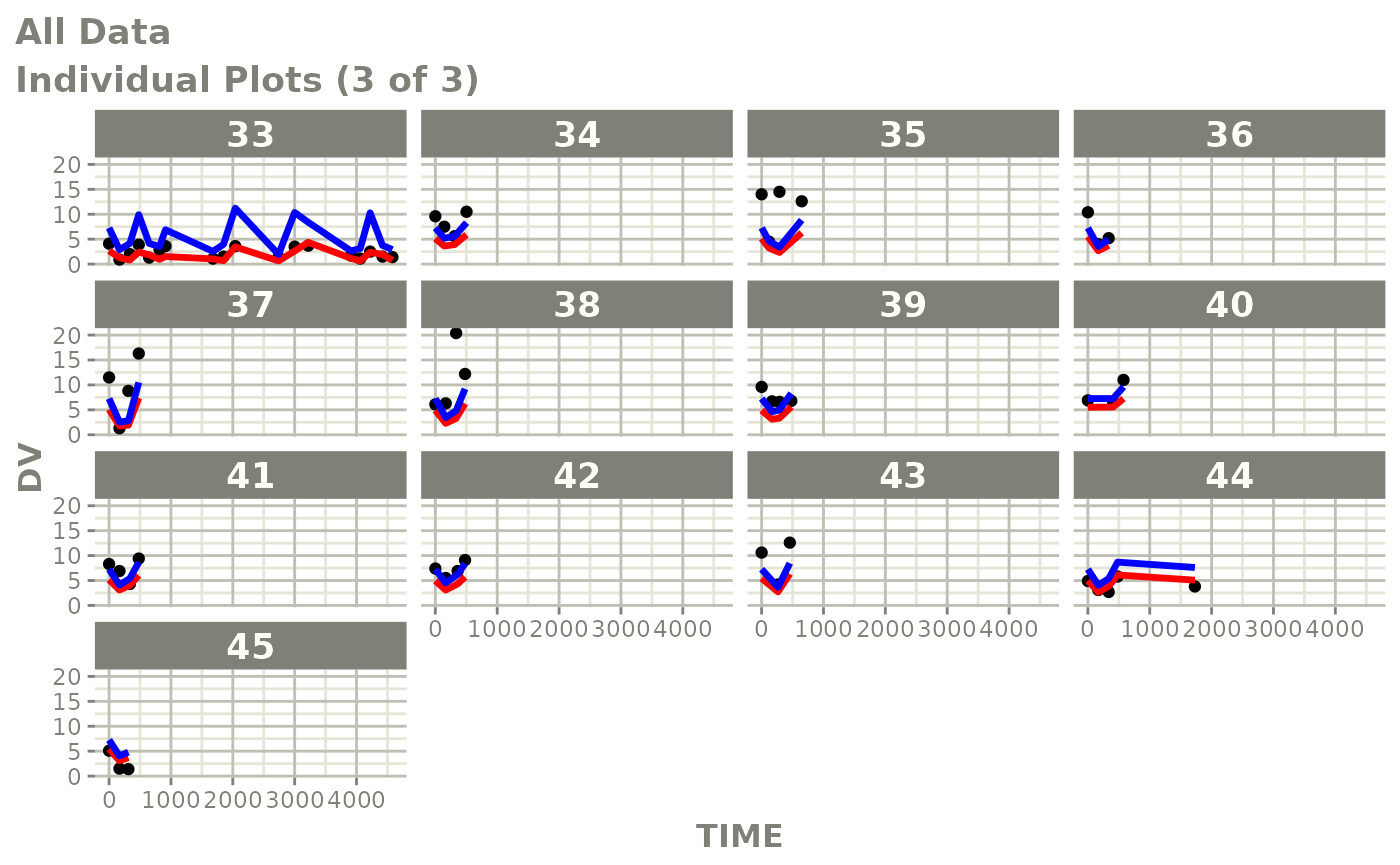
print(dv_vs_pred(xpdb) +
ylab("Observed Neutrophil Count (10^9/L)") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))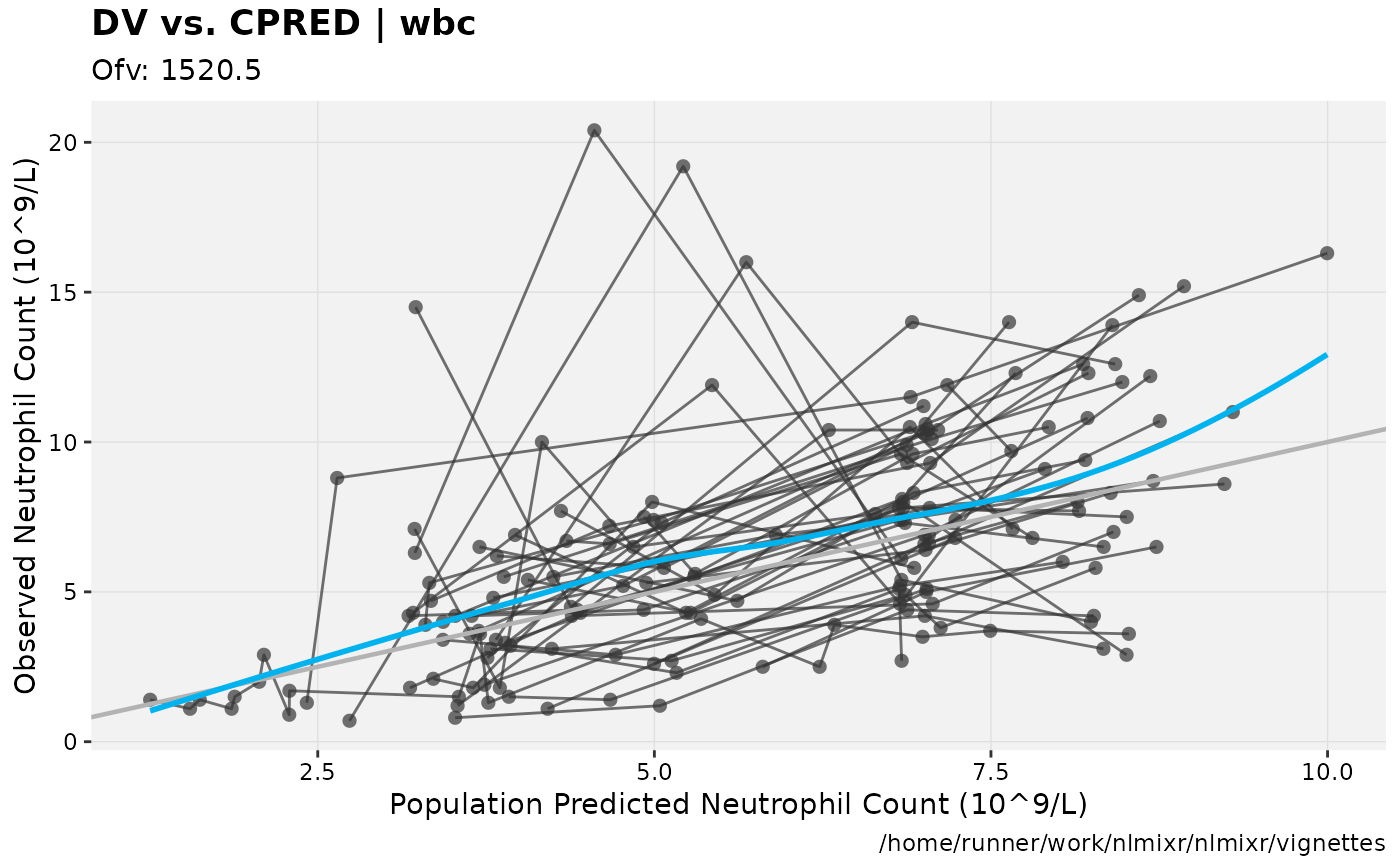
print(dv_vs_ipred(xpdb) +
ylab("Observed Neutrophil Count (10^9/L)") +
xlab("Individual Predicted Neutrophil Count (10^9/L)"))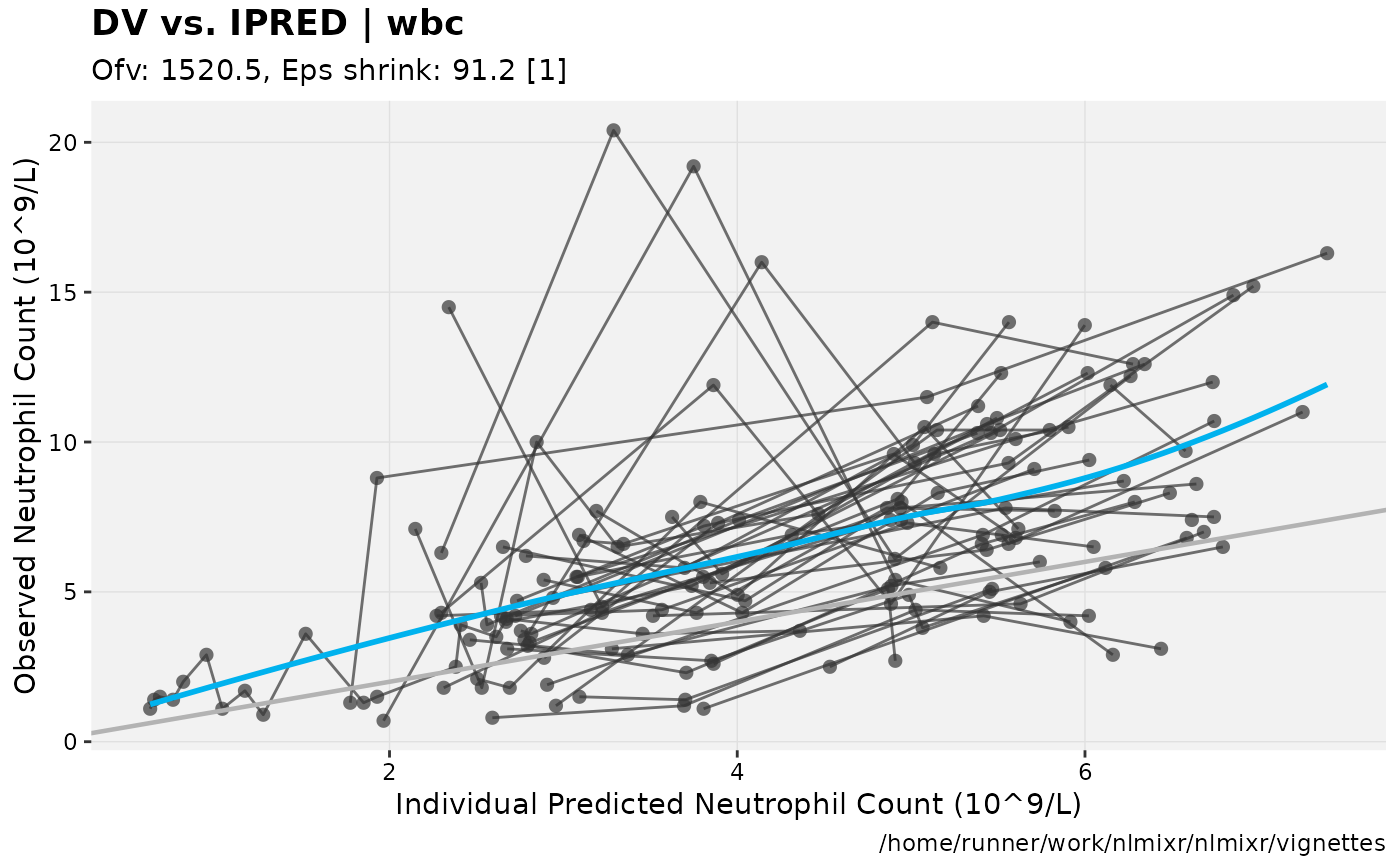
print(res_vs_pred(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))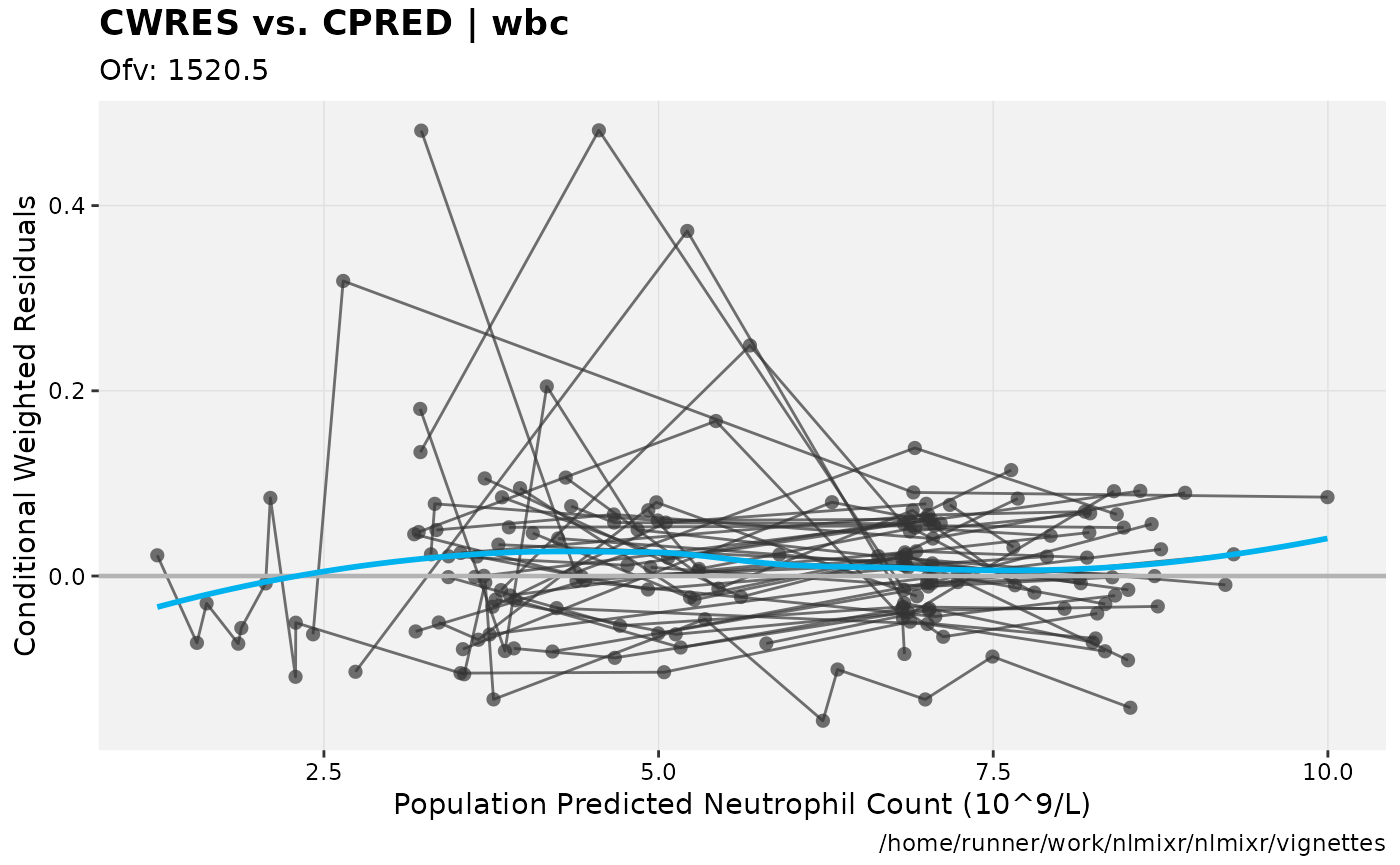
print(res_vs_idv(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Time (h)"))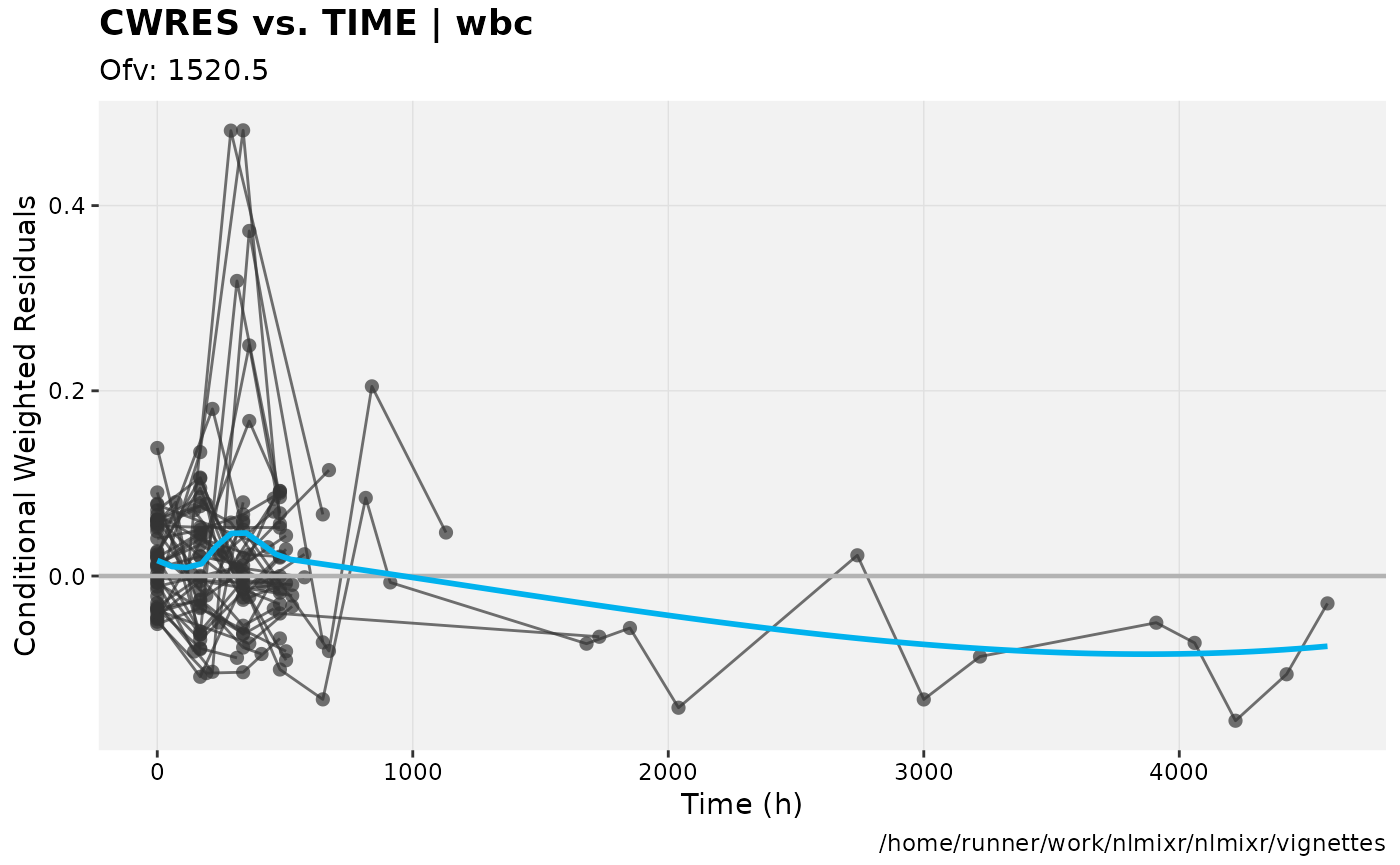
print(absval_res_vs_idv(xpdb, res = 'IWRES') +
ylab("Individual Weighted Residuals") +
xlab("Time (h)"))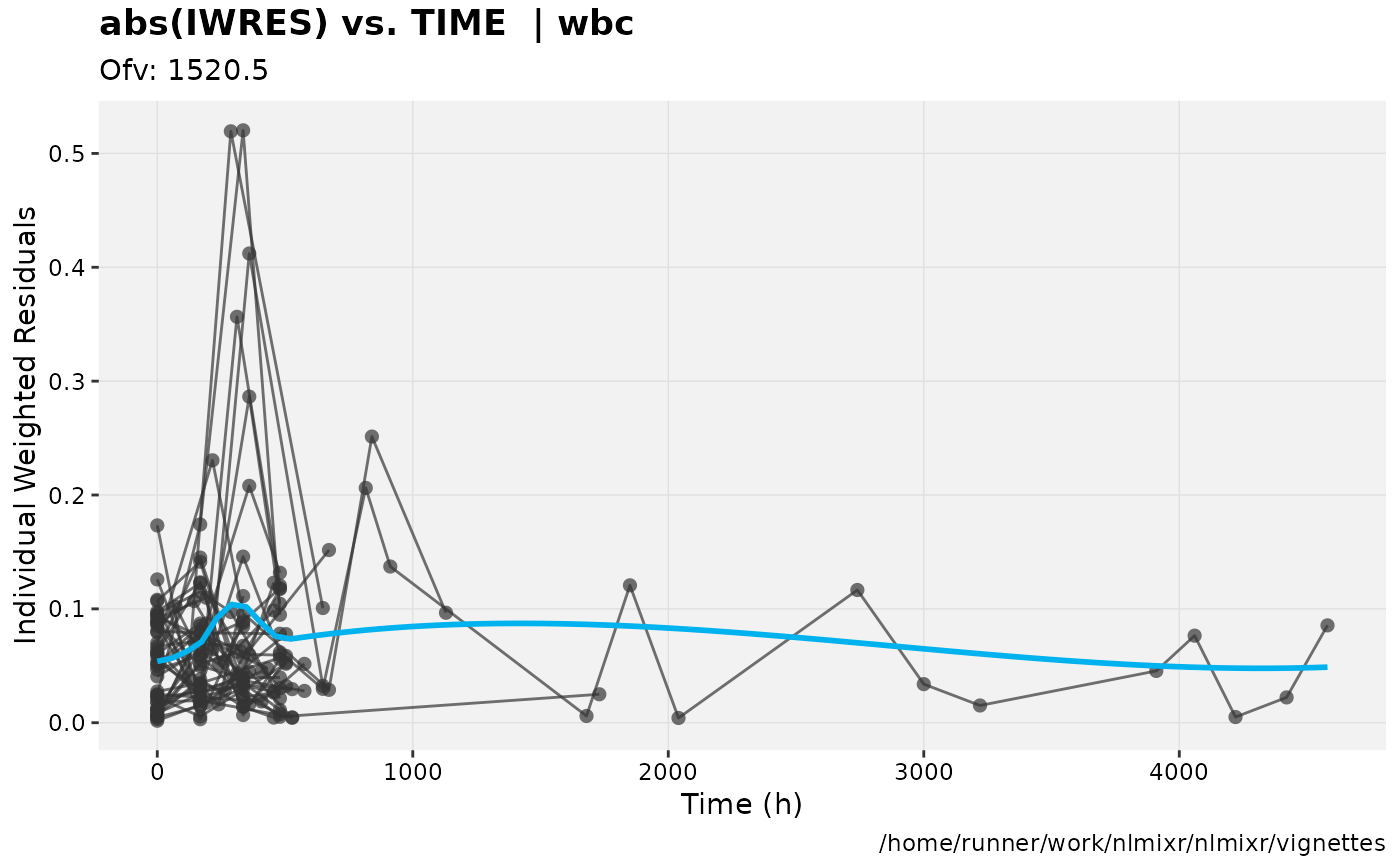
print(absval_res_vs_pred(xpdb, res = 'IWRES') +
ylab("Individual Weighted Residuals") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))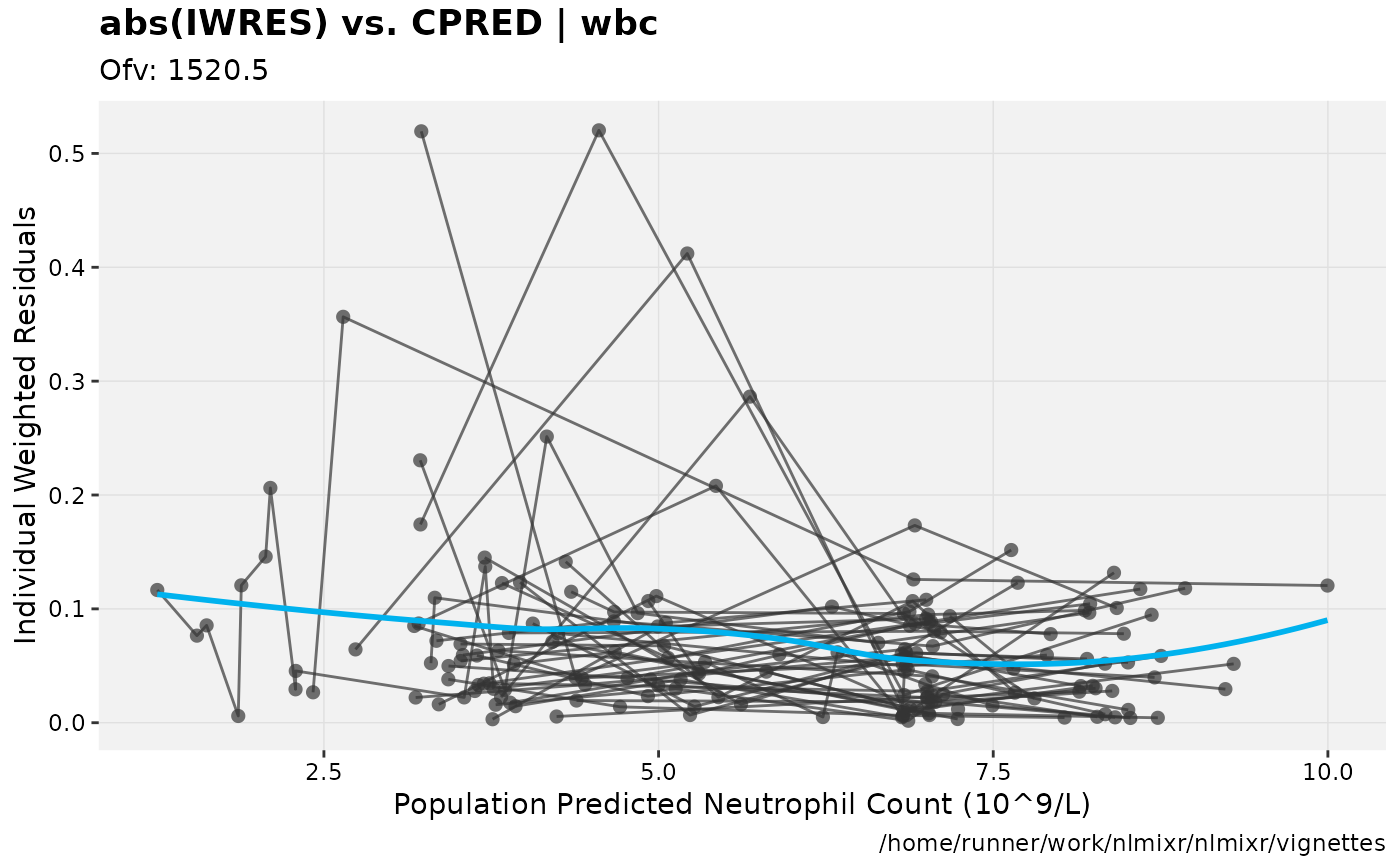
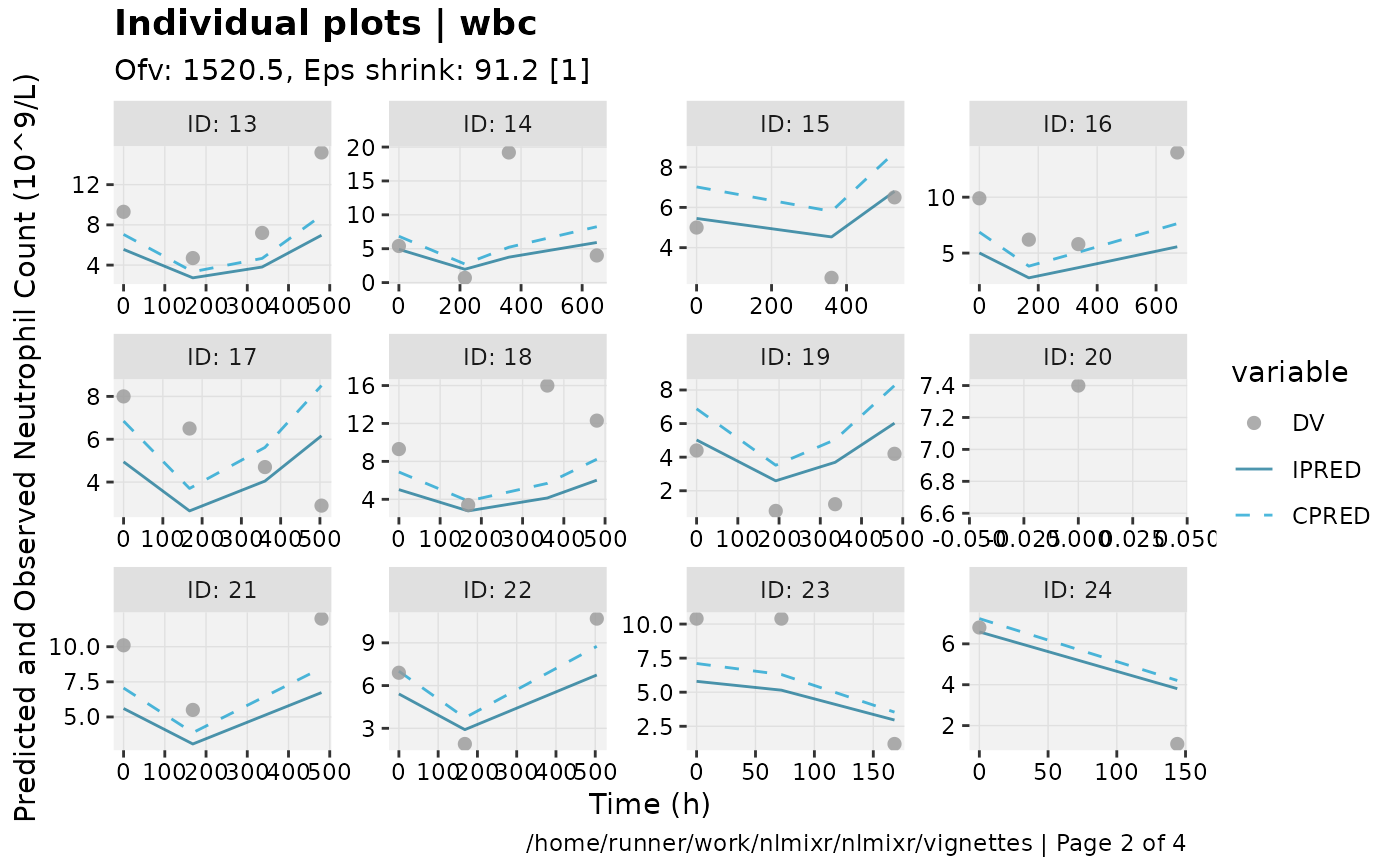
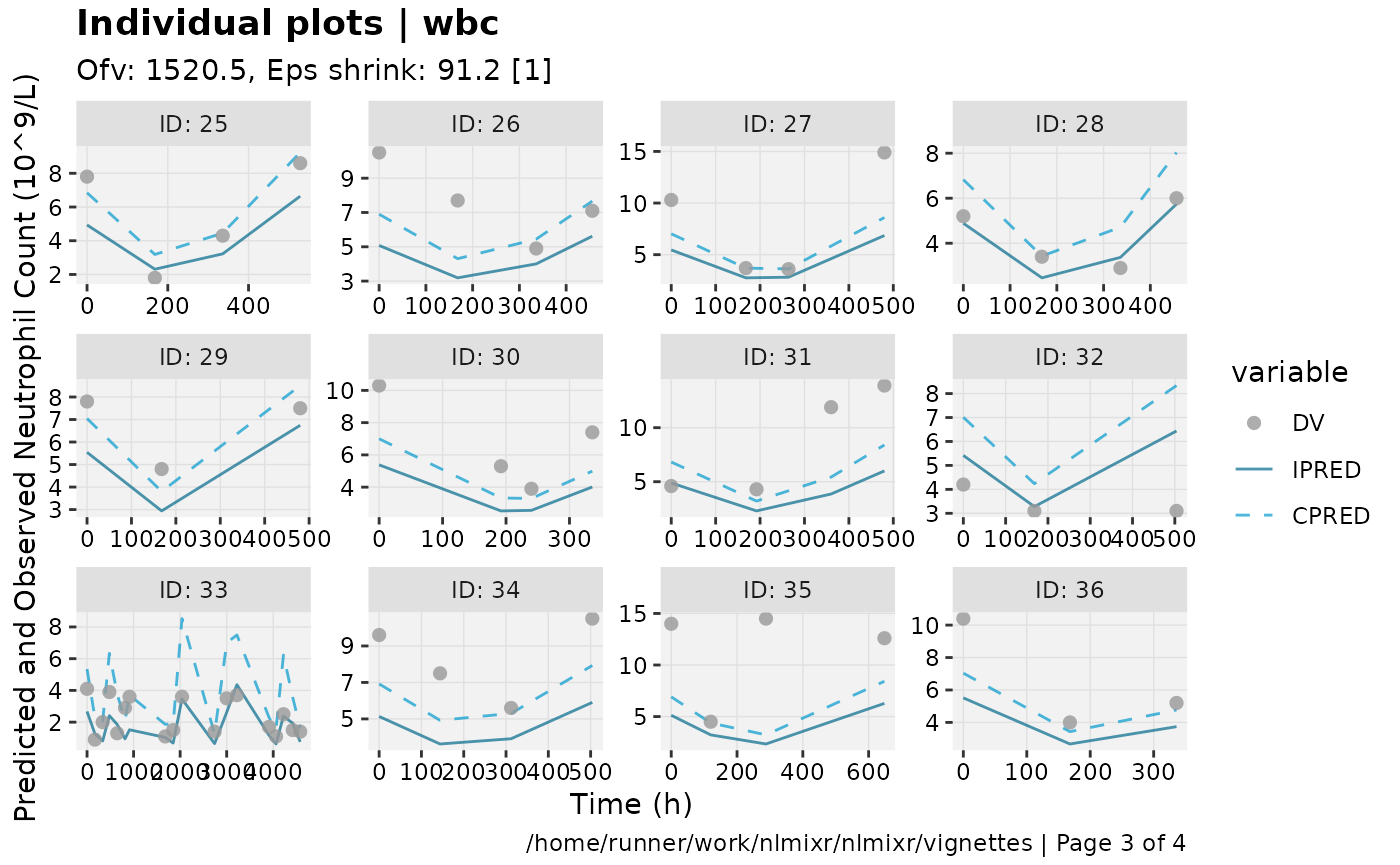
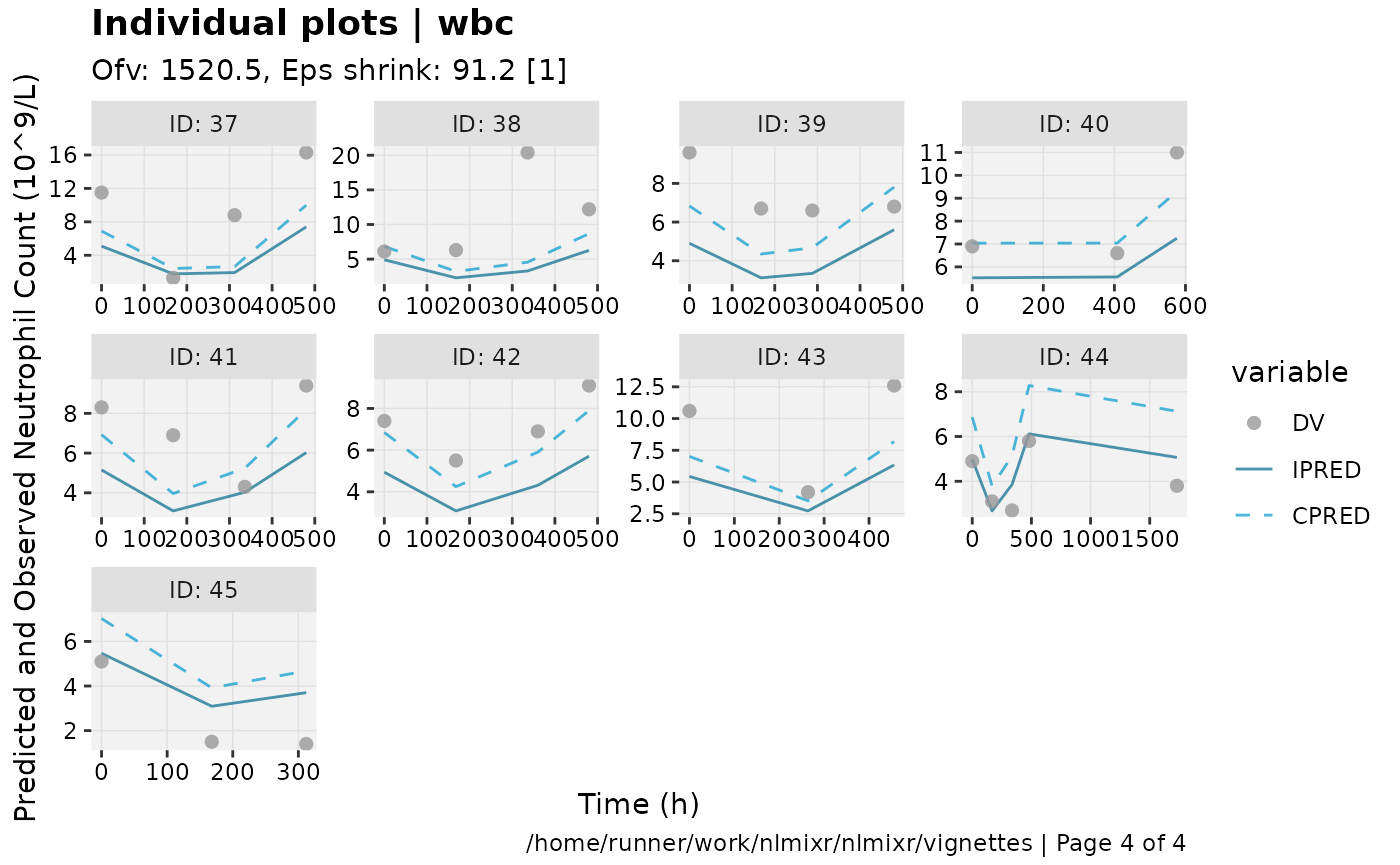
print(ind_plots(xpdb, nrow=3, ncol=4) +
ylab("Predicted and Observed Neutrophil Count (10^9/L)") +
xlab("Time (h)"))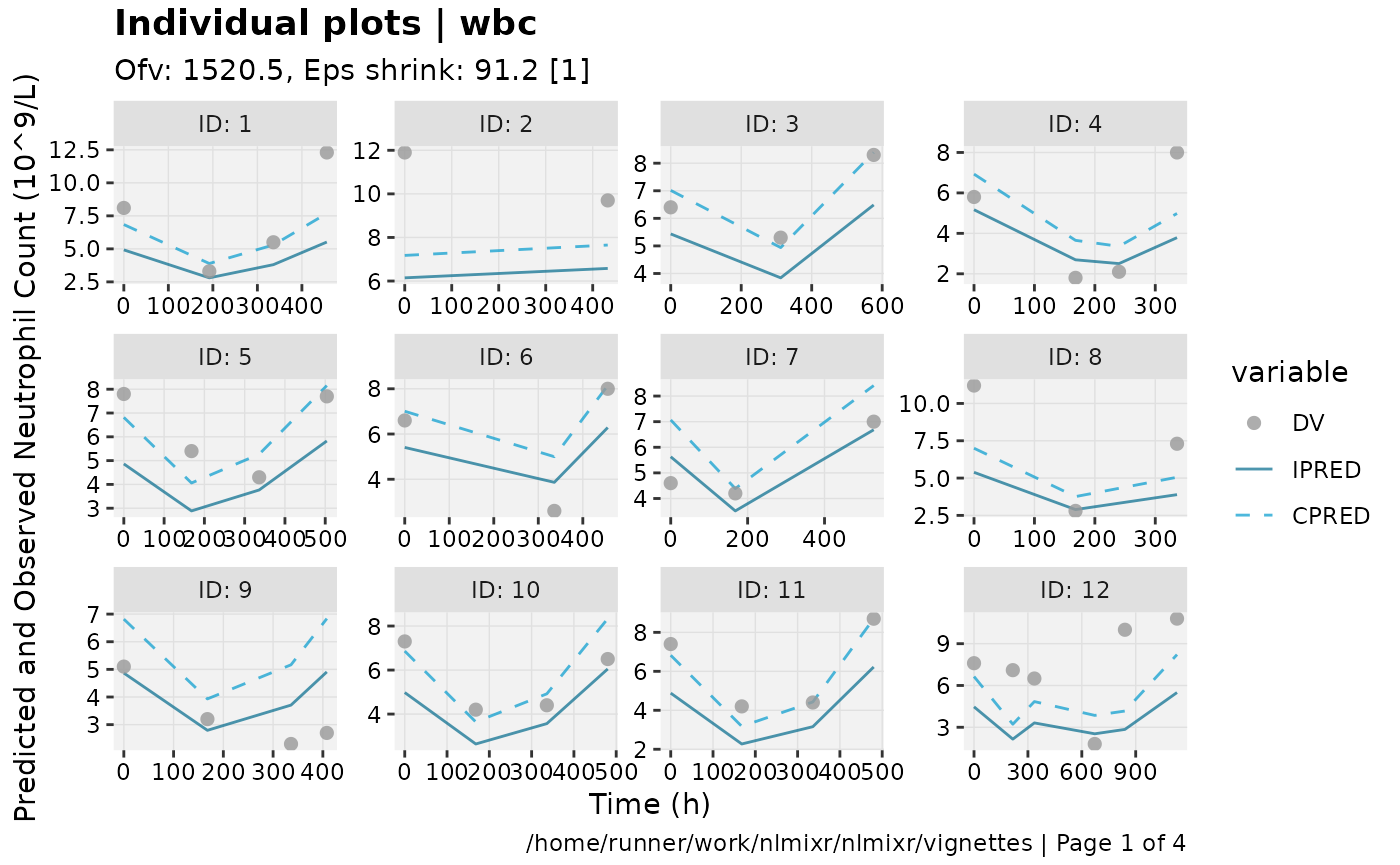
print(res_distrib(xpdb) +
ylab("Density") +
xlab("Conditional Weighted Residuals"))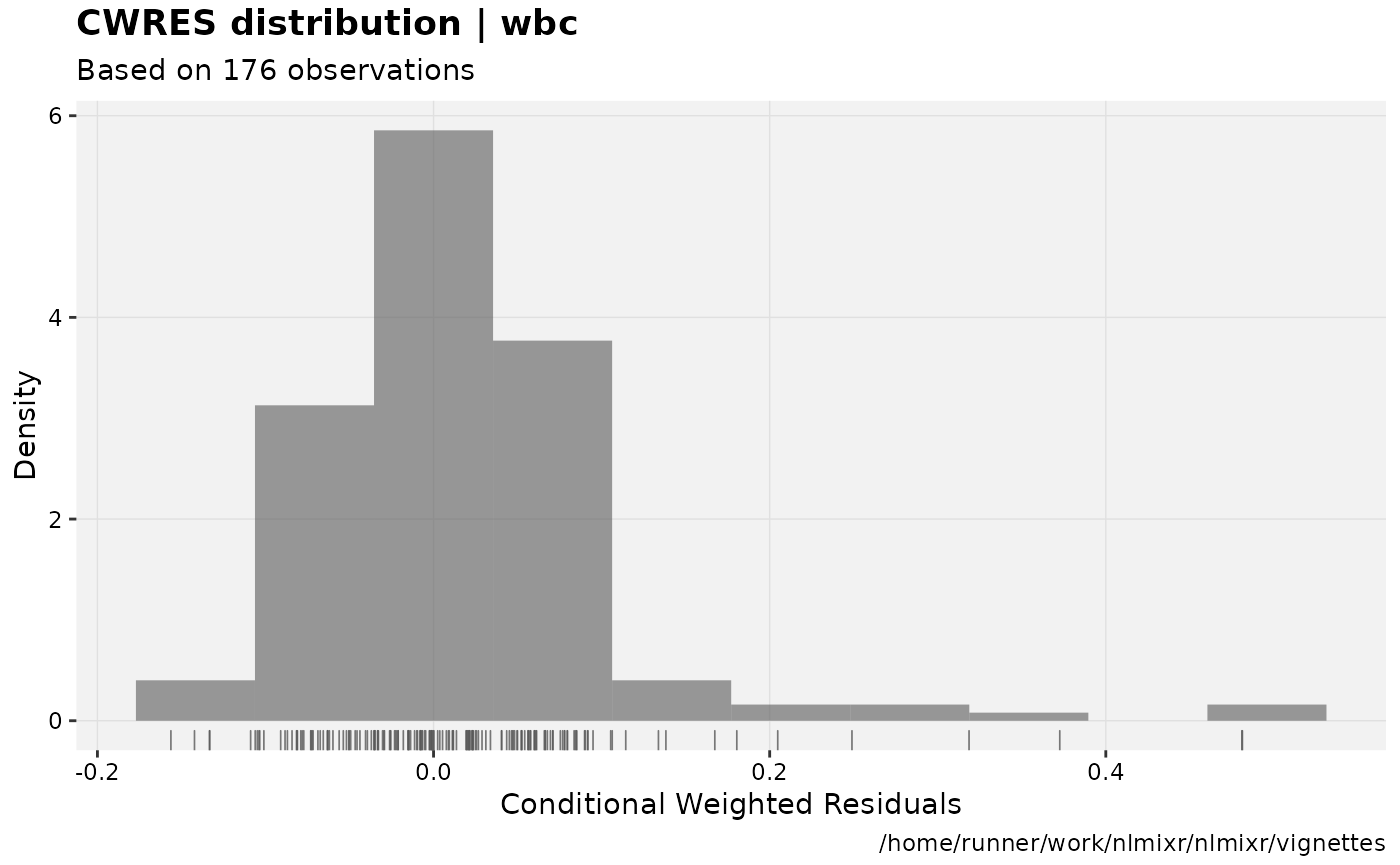
#vpc.ui(fit.S, n=500, show=list(obs_dv=T), ylab = "Neutrophil count (10^9/L)", xlab = "Time (h)")
# 10 bins is slightly better than auto bin
vpc.ui(fit.F, n=500, n_bins = 10, show=list(obs_dv=T), ylab = "Neutrophil Count (10^9/L)", xlab = "Time (h)")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:05
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now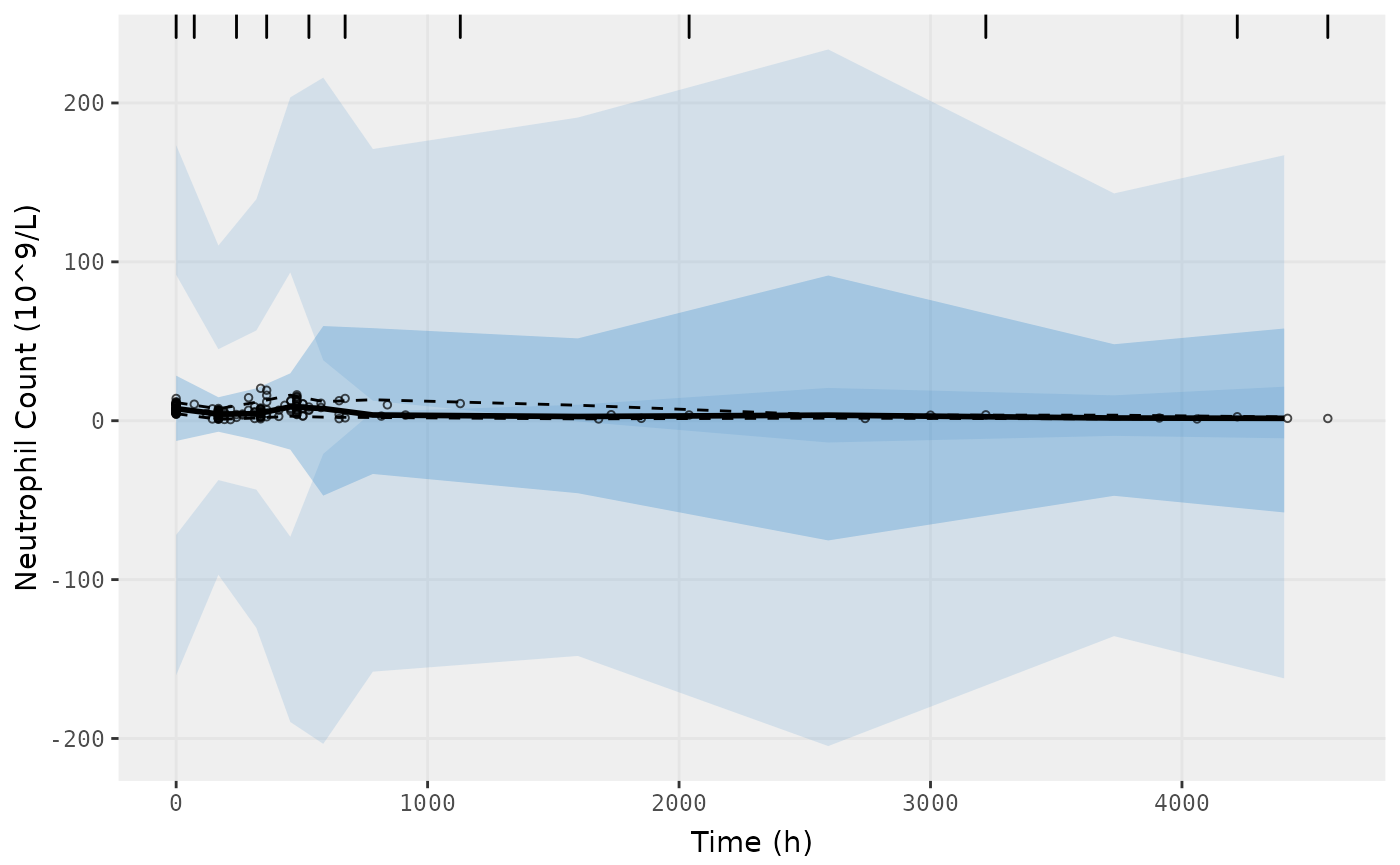
# specify bins
vpc.ui(fit.F, n=500, bins = c(0,170,300,350,500,600,900,3000,4580), show=list(obs_dv=T), ylab = "Neutrophil Count (10^9/L)", xlab = "Time (h)")
#> [====|====|====|====|====|====|====|====|====|====] 0:00:05
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now