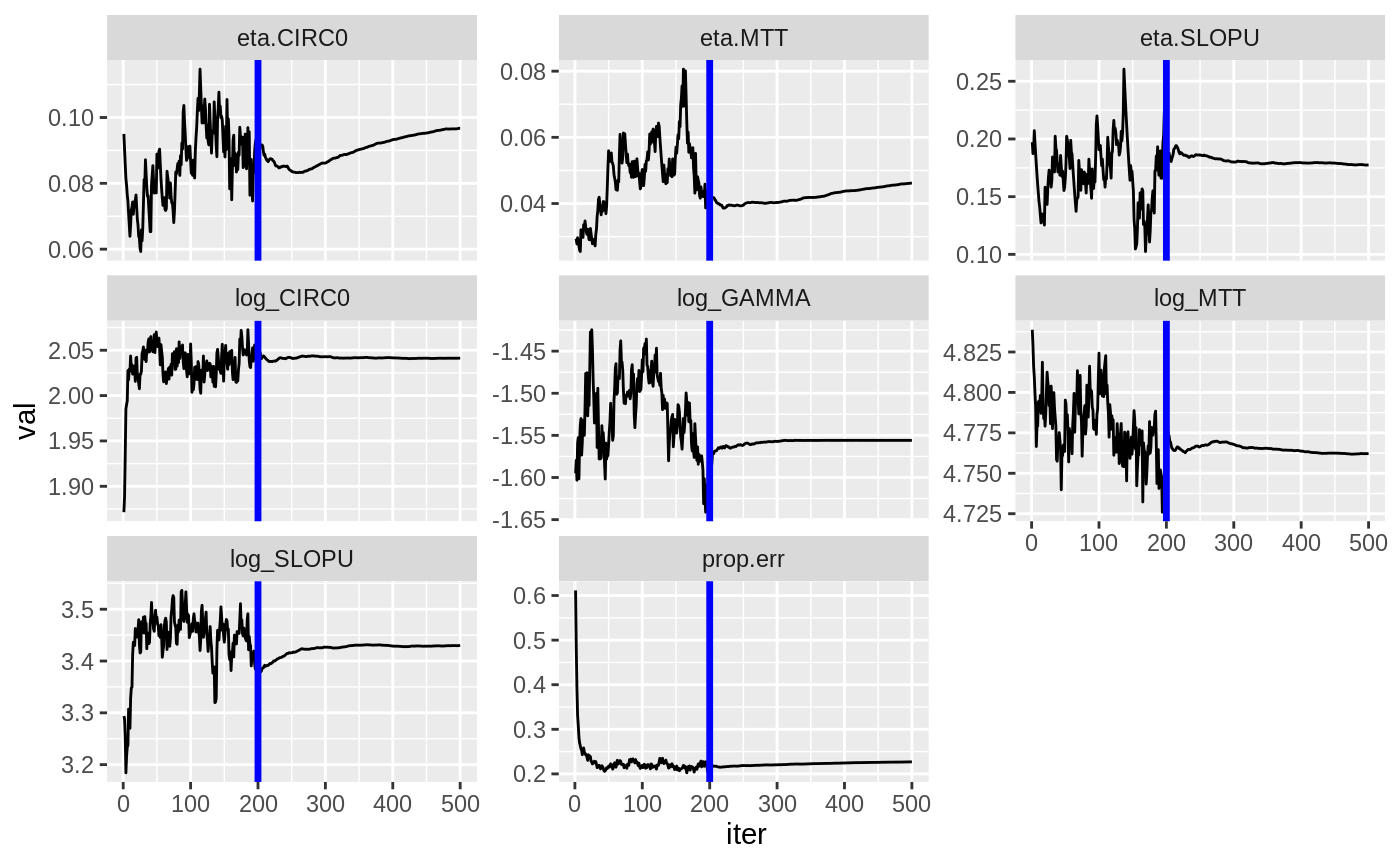
Fit model using saem
d3 = read.csv("Simulated_WBC_pacl_ddmore_samePK_nlmixr.csv", na.string = ".")
fit.S <- nlmixr(wbc, d3, est="saem", table=list(cwres=TRUE, npde=TRUE))
#> Loading model already run (/tmp/RtmpXKdUFj/temp_libpath58be457a881a/nlmixr.examples/nlmixr-wbc-d3-saem-f1f1b2aae1f74678ea021385c3b62987.rds)
#> Warning in lapply(X = X, FUN = FUN, ...): ID missing in parameters dataset;
#> Parameters are assumed to have the same order as the IDs in the event dataset
xpdb <- xpose_data_nlmixr(fit.S)
#> Warning: `complete_()` is deprecated as of tidyr 1.0.0.
#> Please use `complete()` instead.
#> This warning is displayed once per session.
#> Call `lifecycle::last_warnings()` to see where this warning was generated.
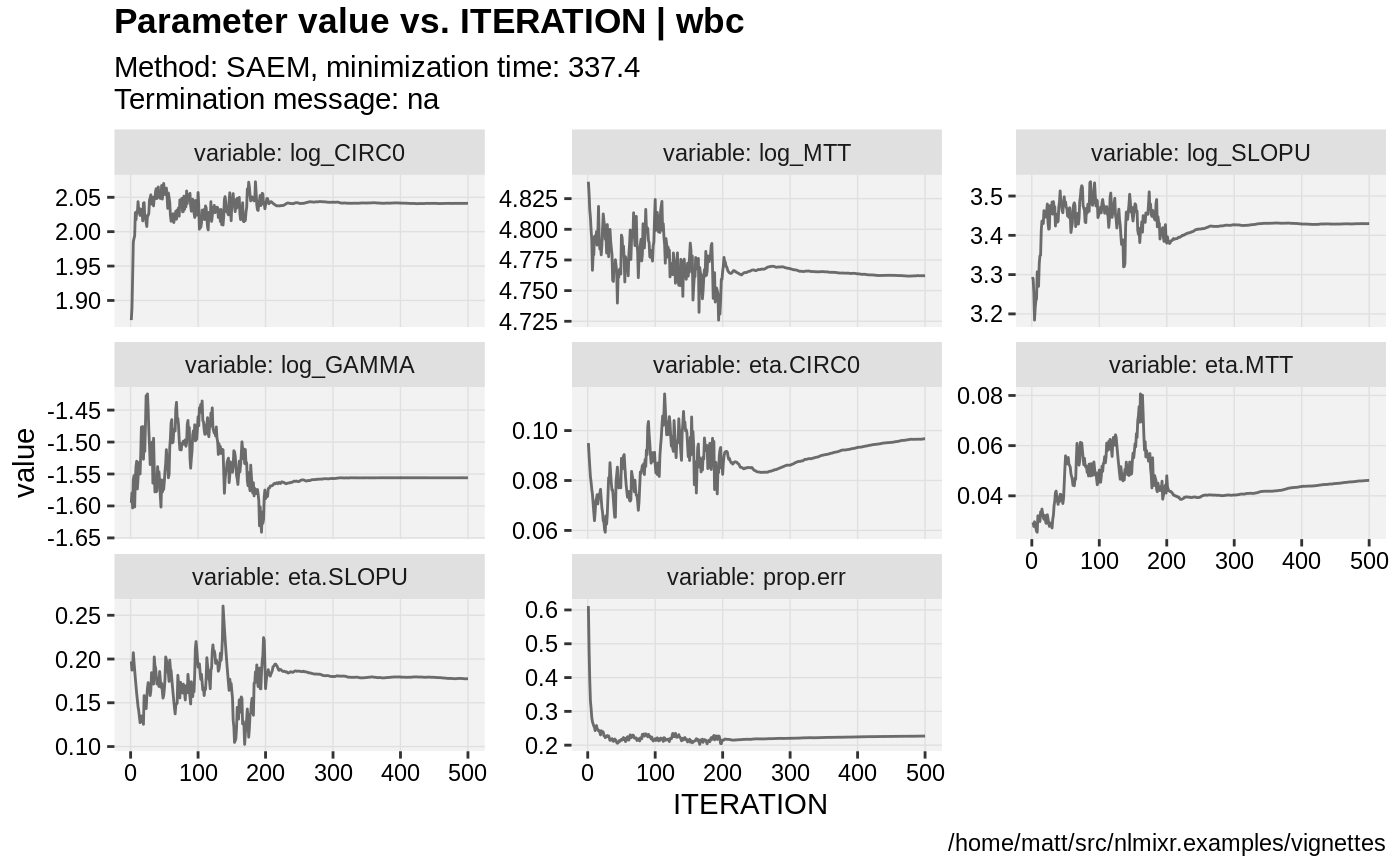
plot(fit.S)
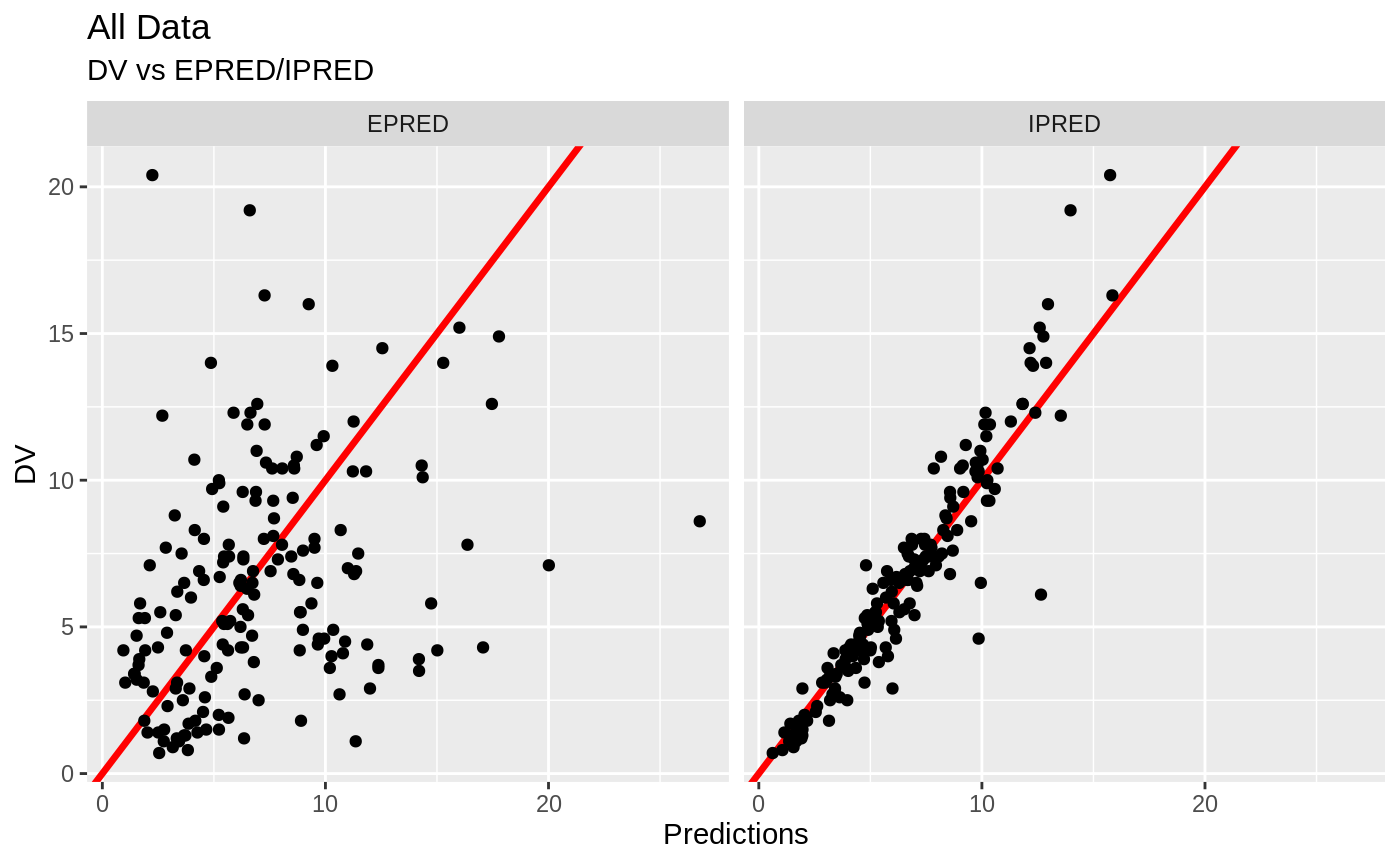
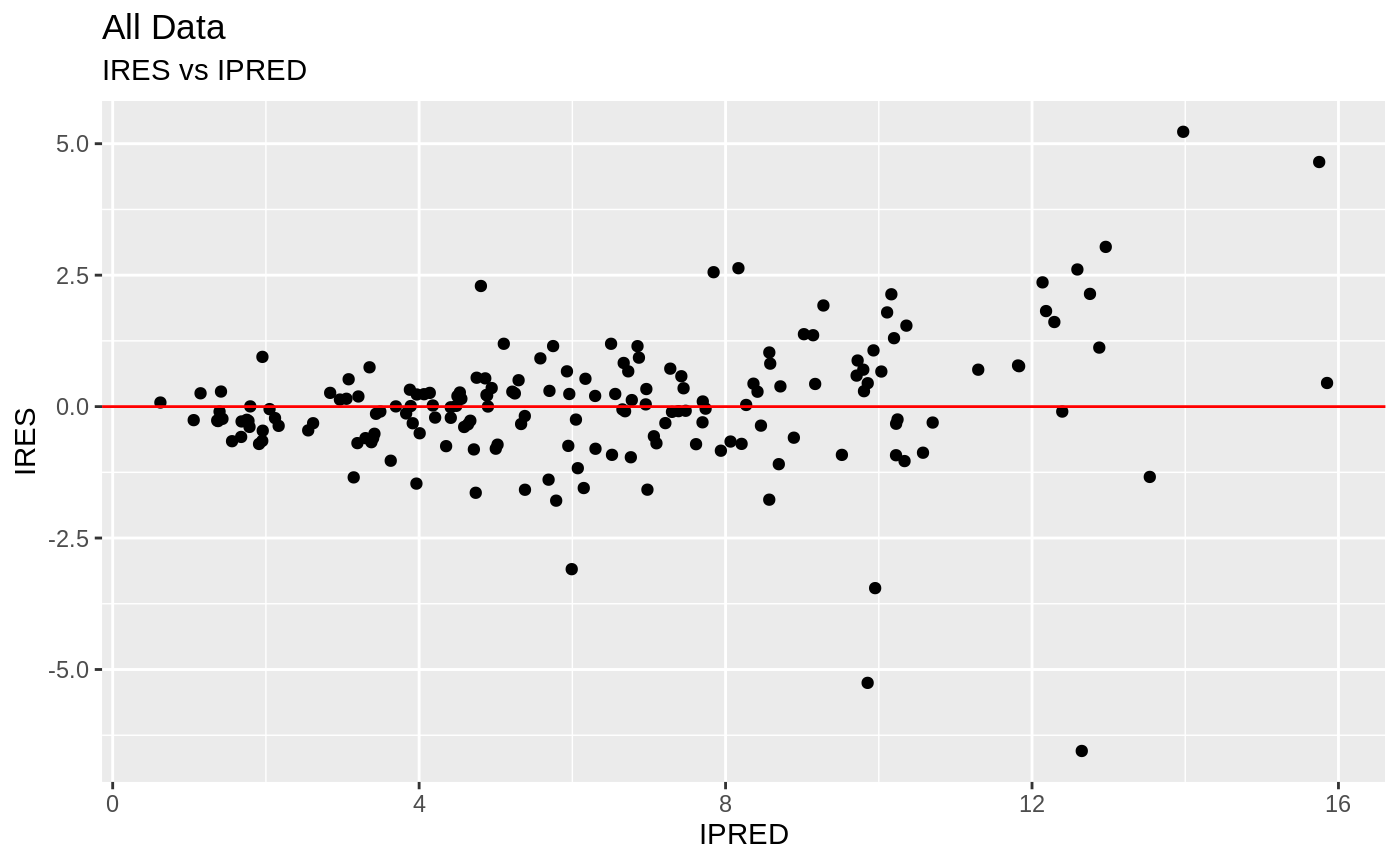
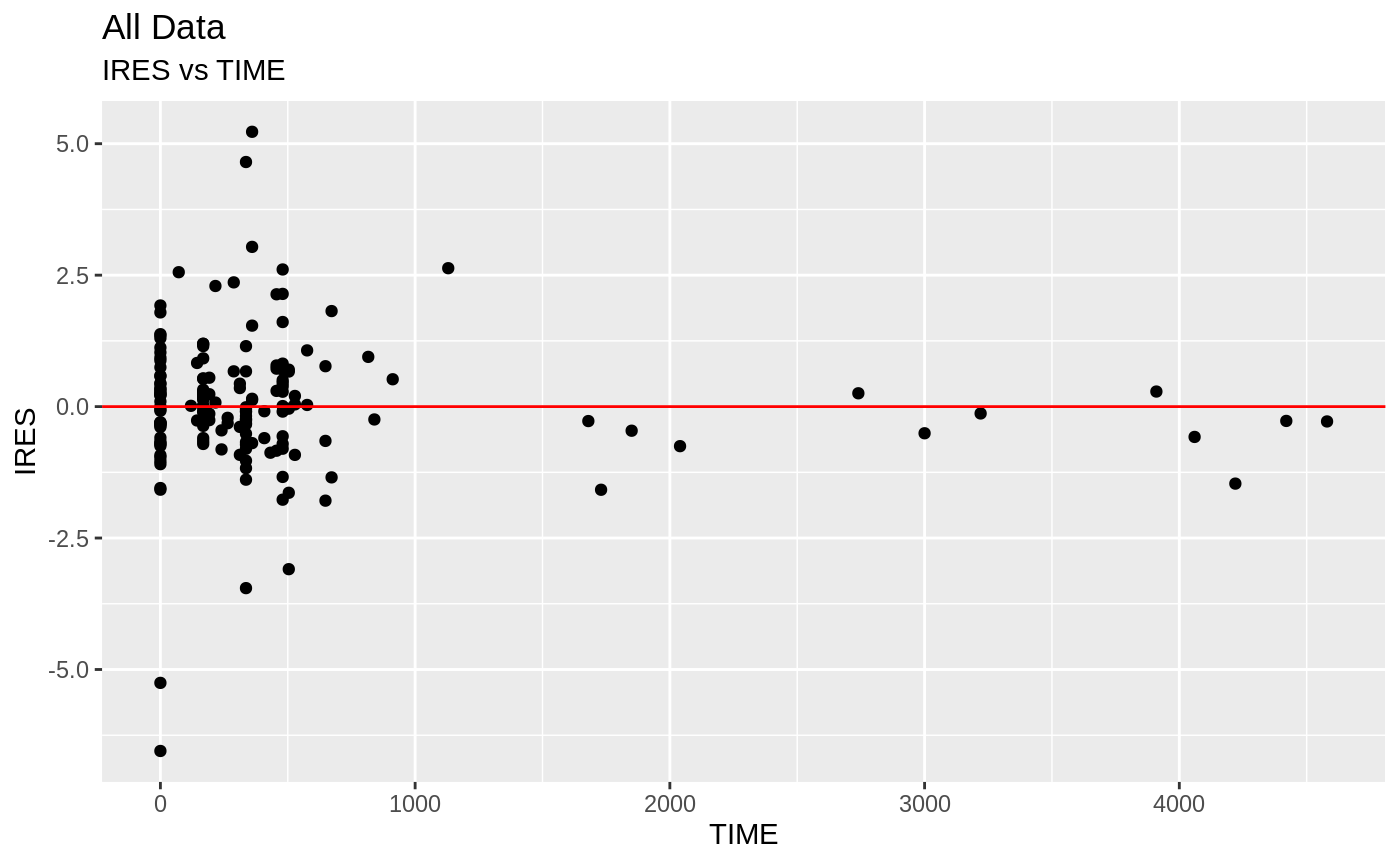
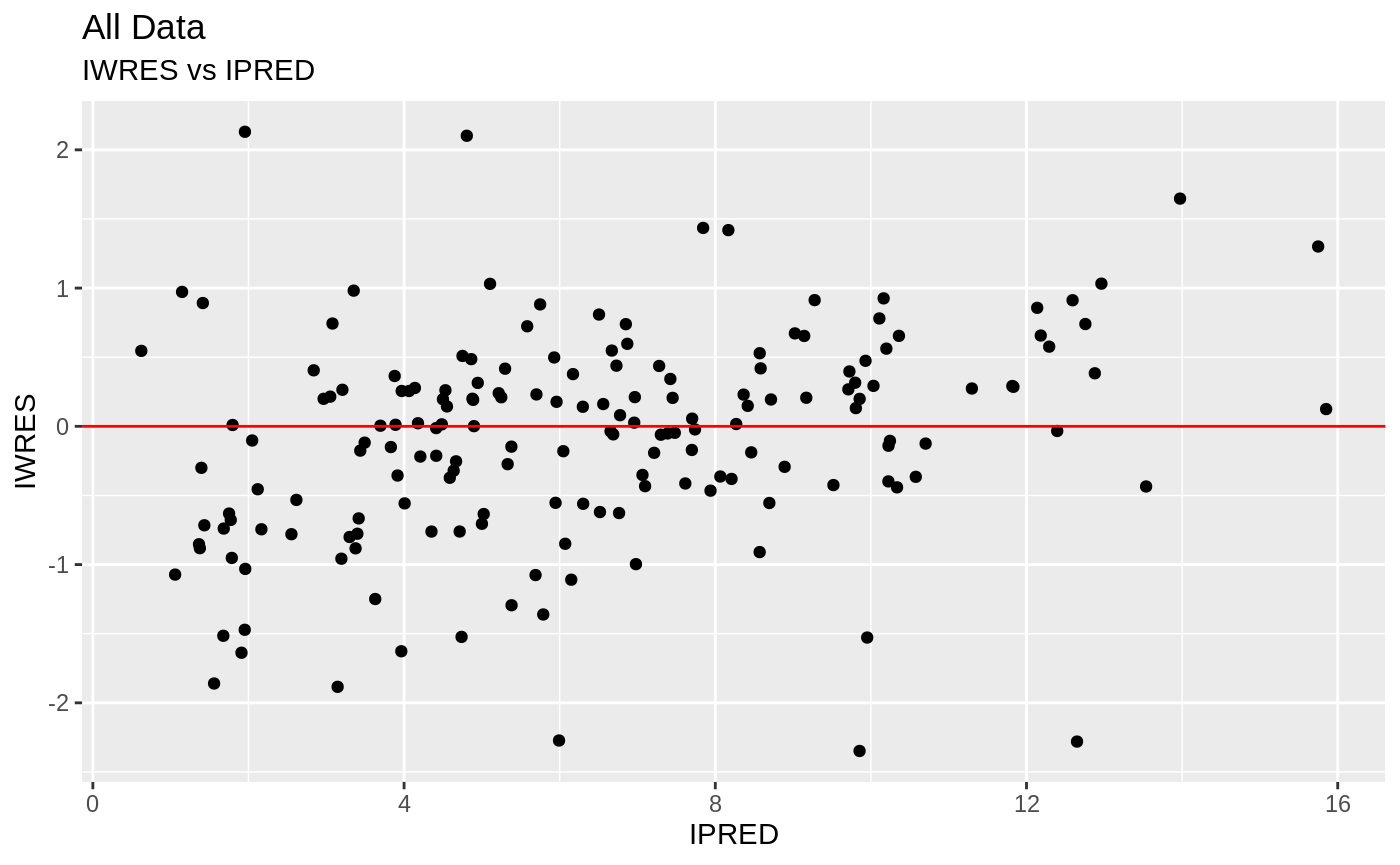
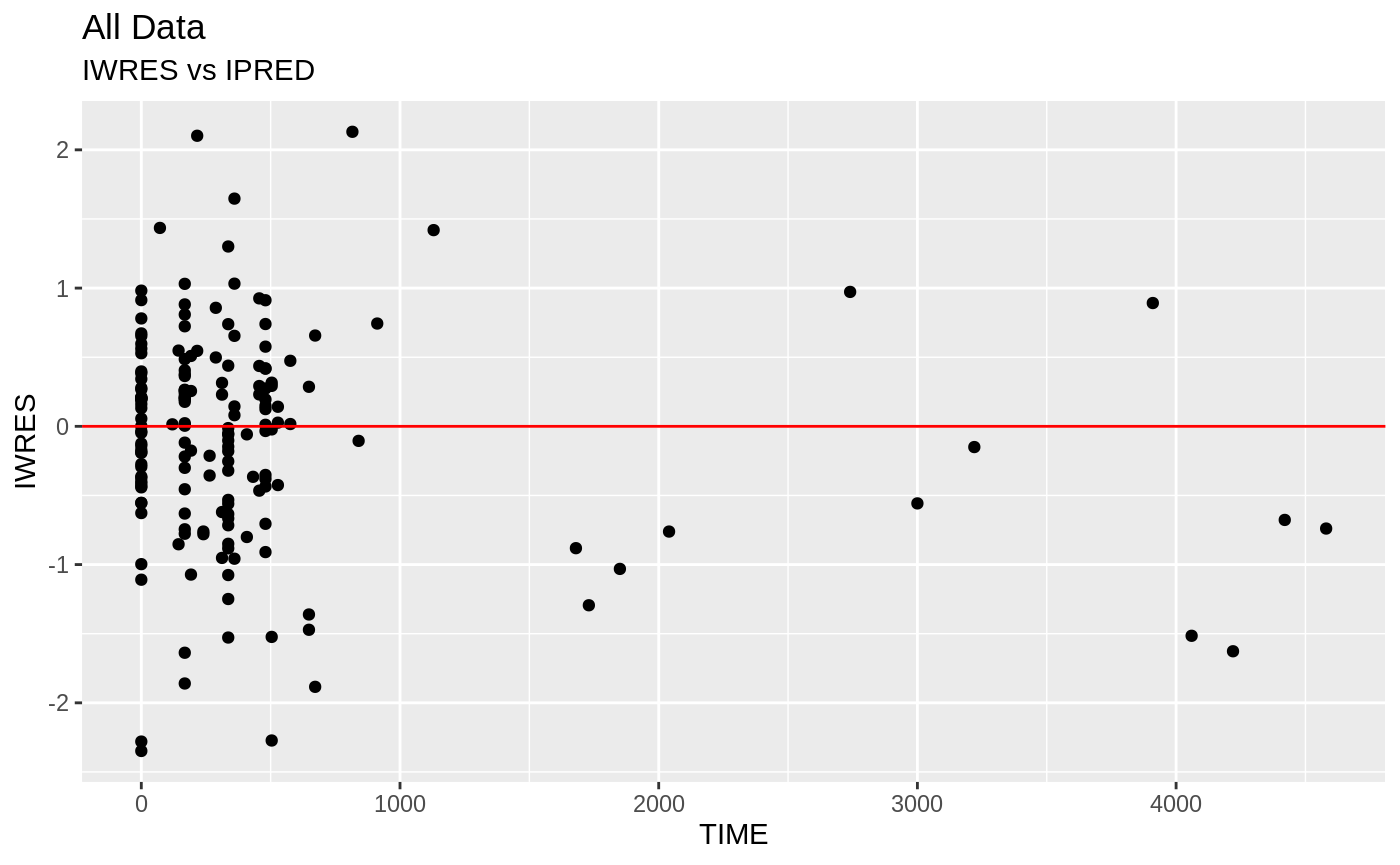
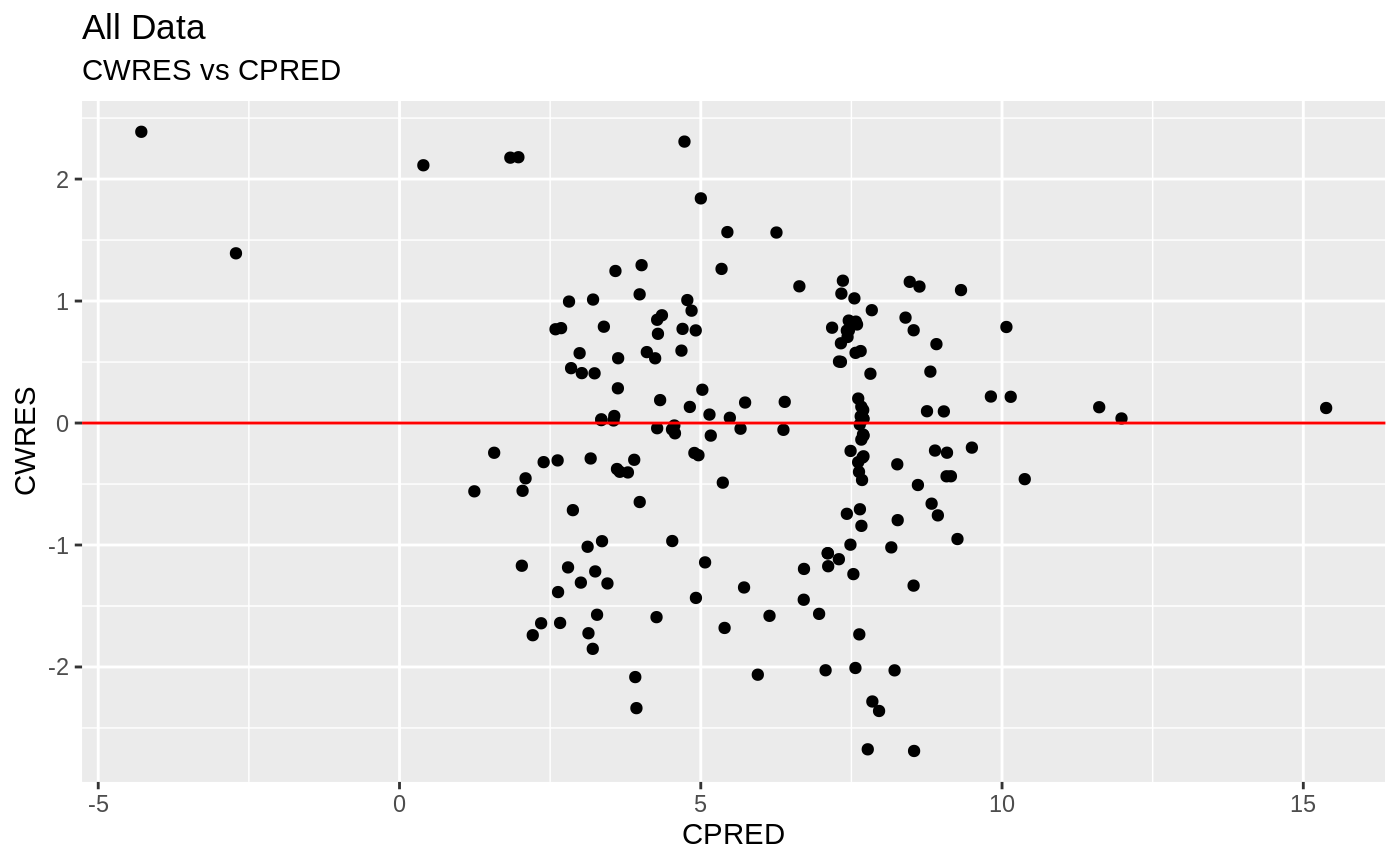
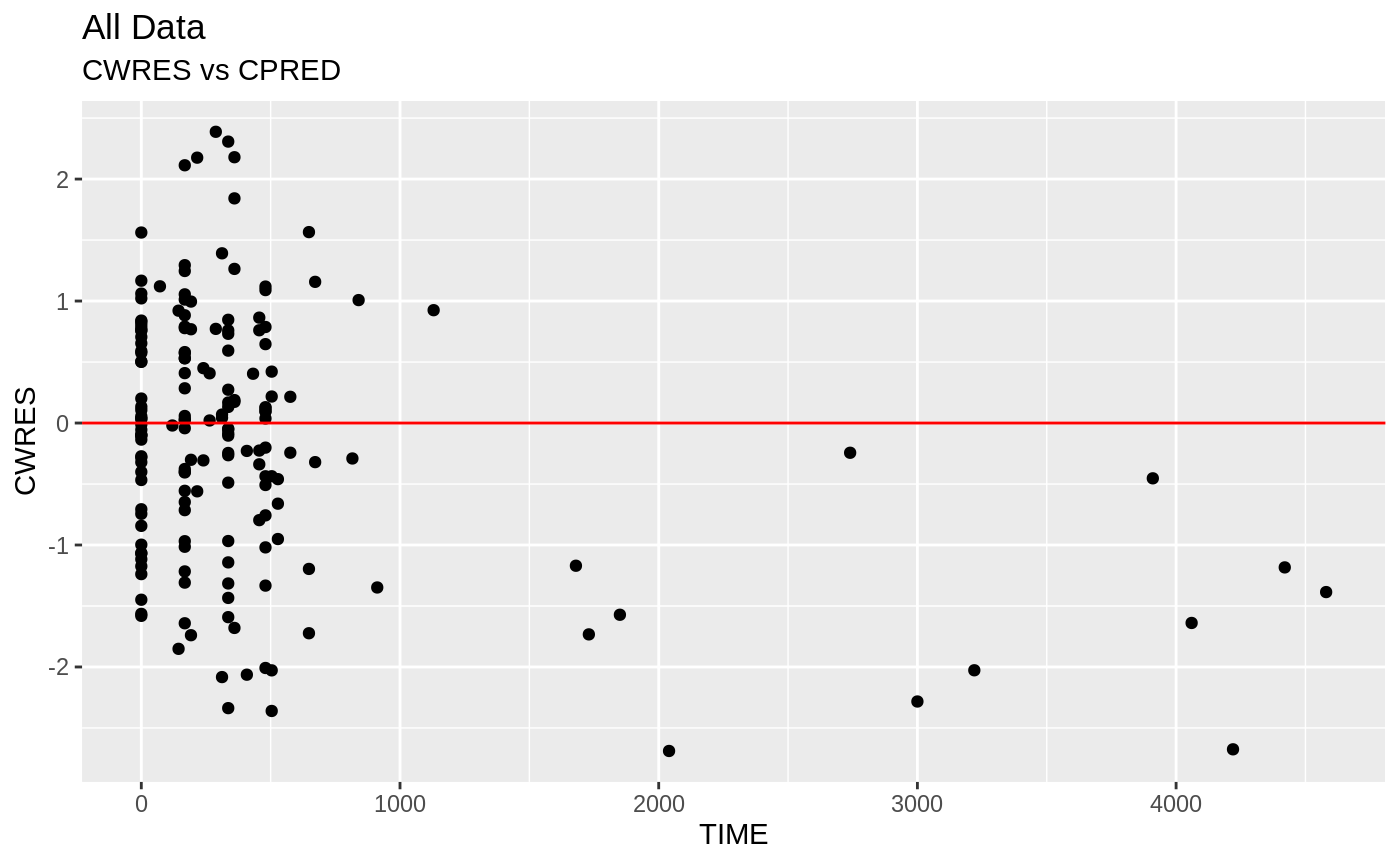
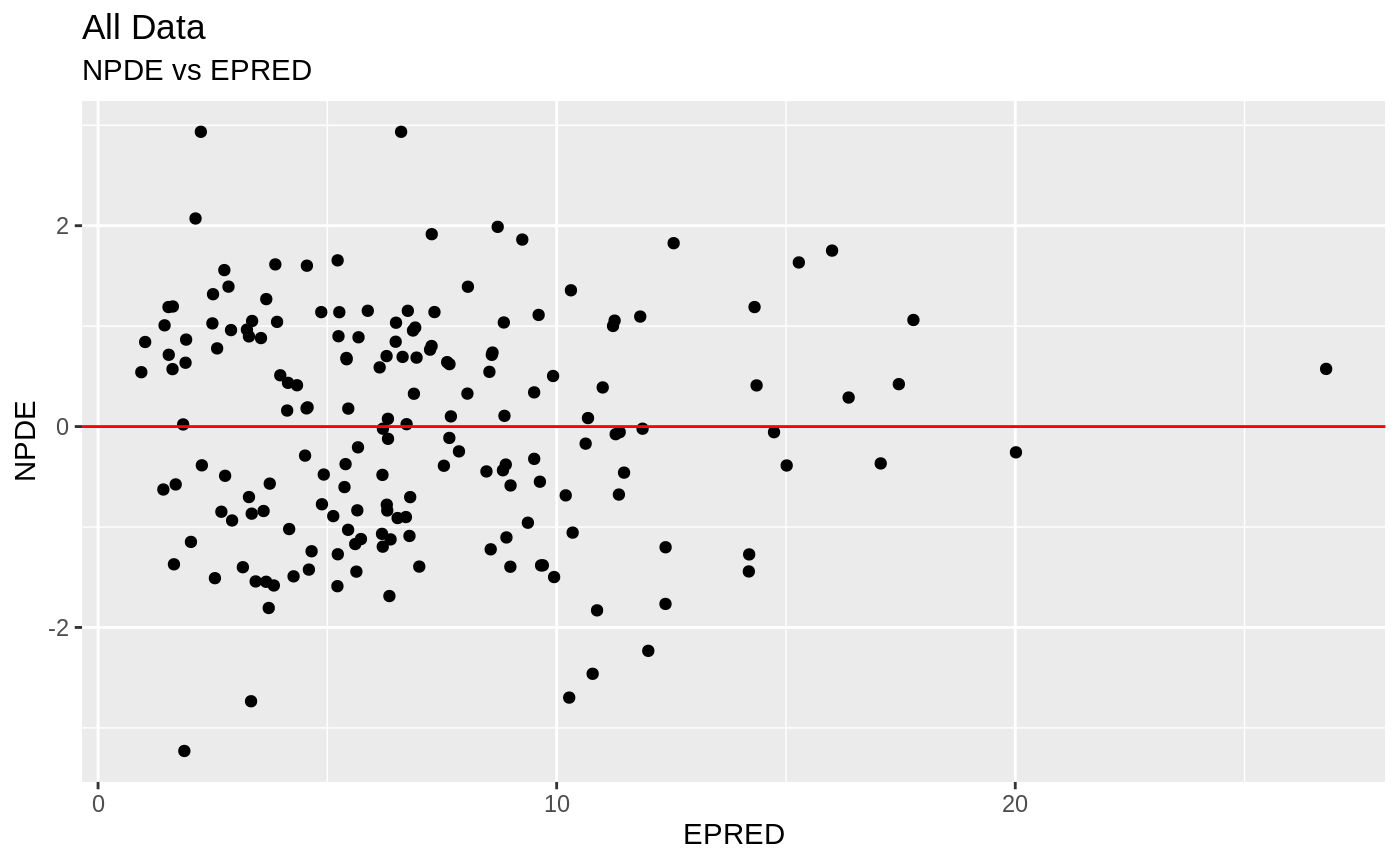
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
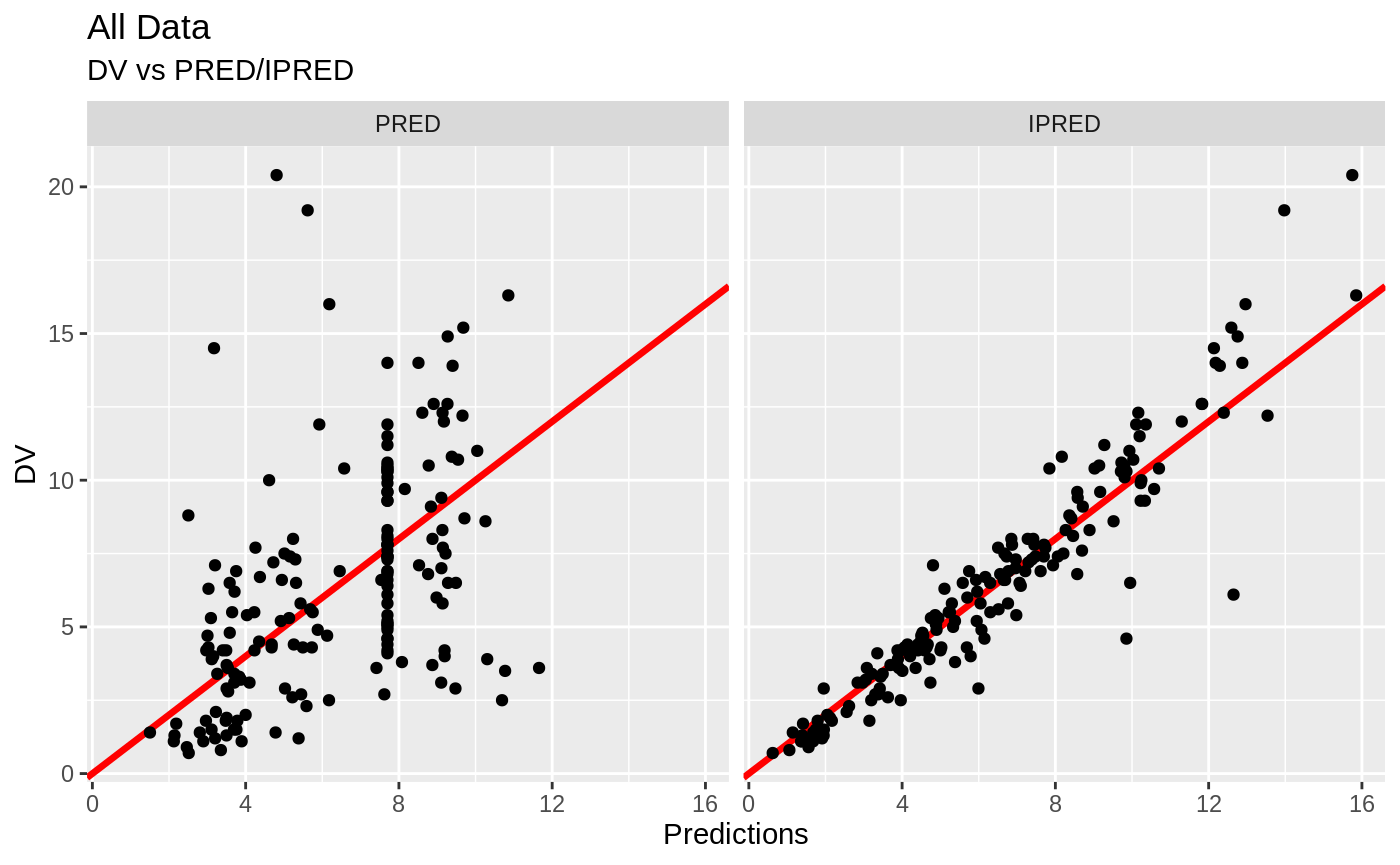
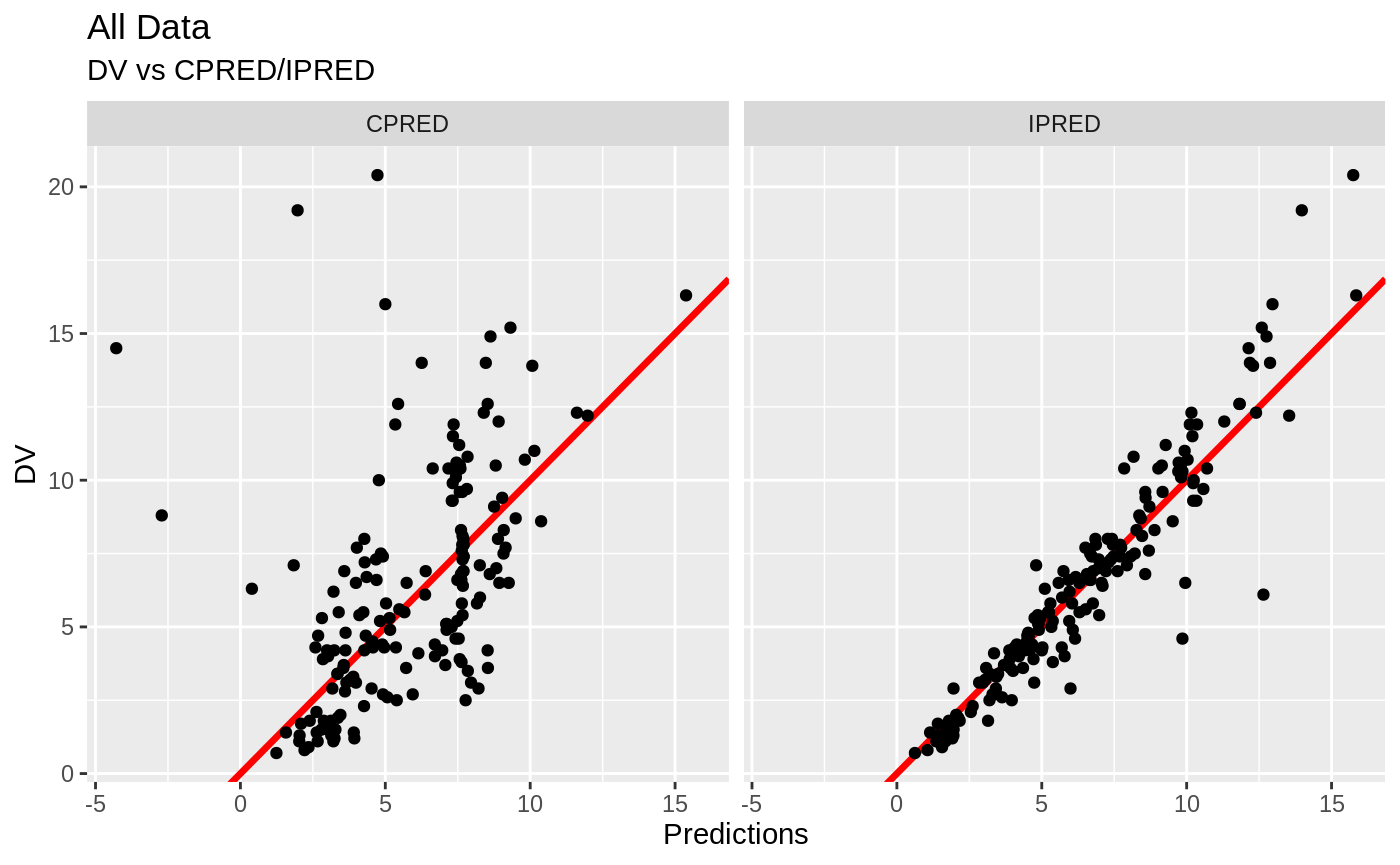
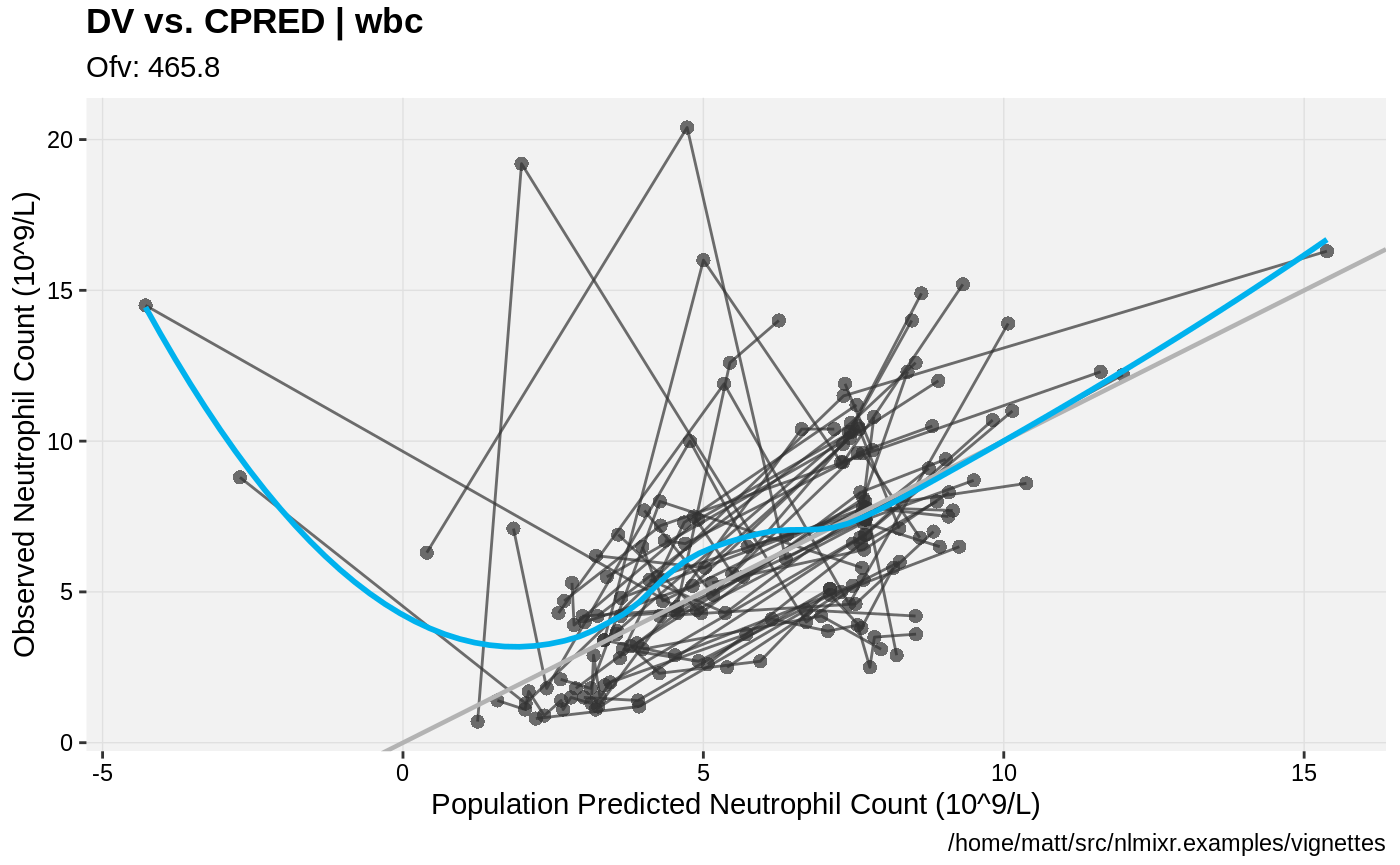
print(dv_vs_pred(xpdb) +
ylab("Observed Neutrophil Count (10^9/L)") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))
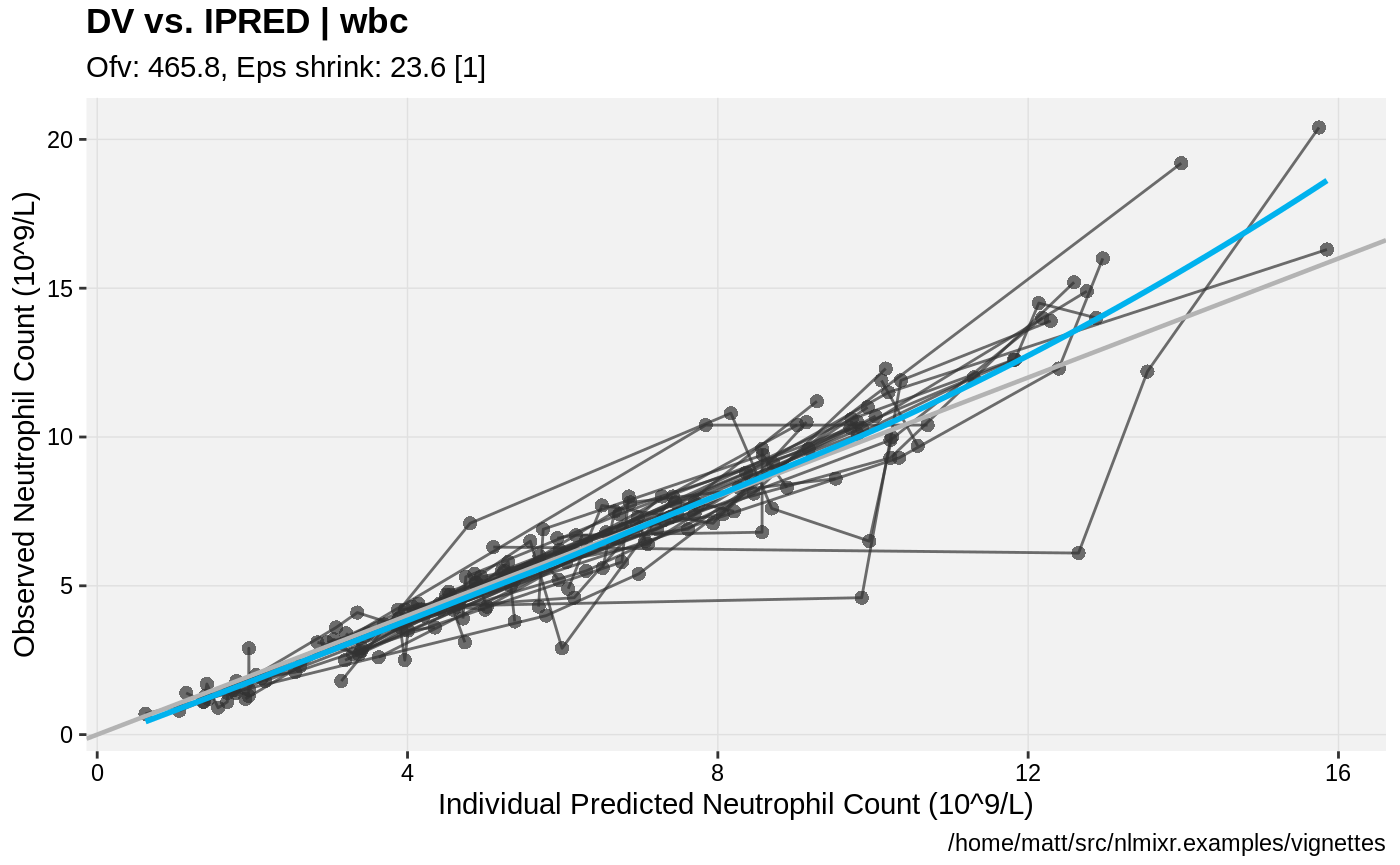
print(dv_vs_ipred(xpdb) +
ylab("Observed Neutrophil Count (10^9/L)") +
xlab("Individual Predicted Neutrophil Count (10^9/L)"))
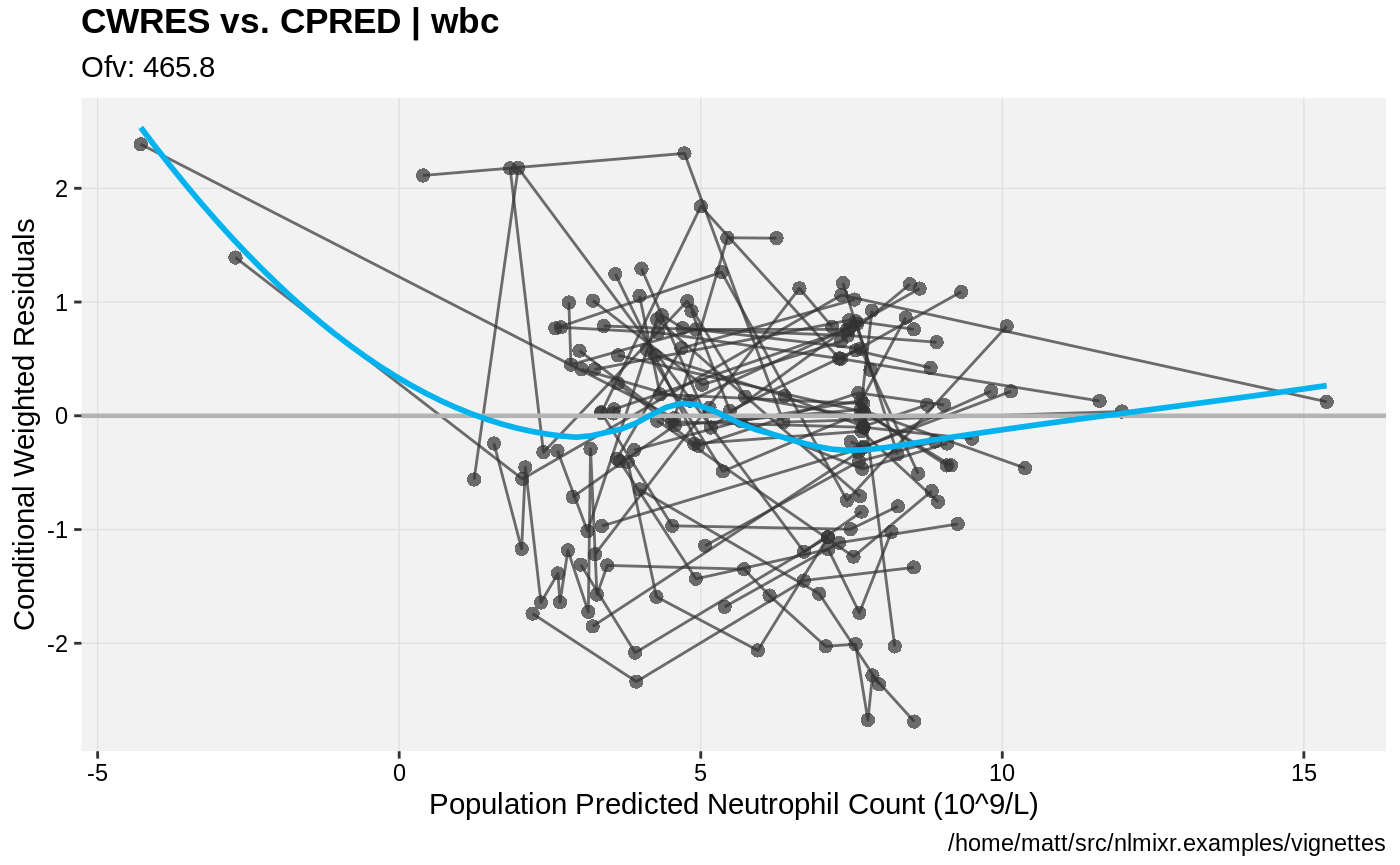
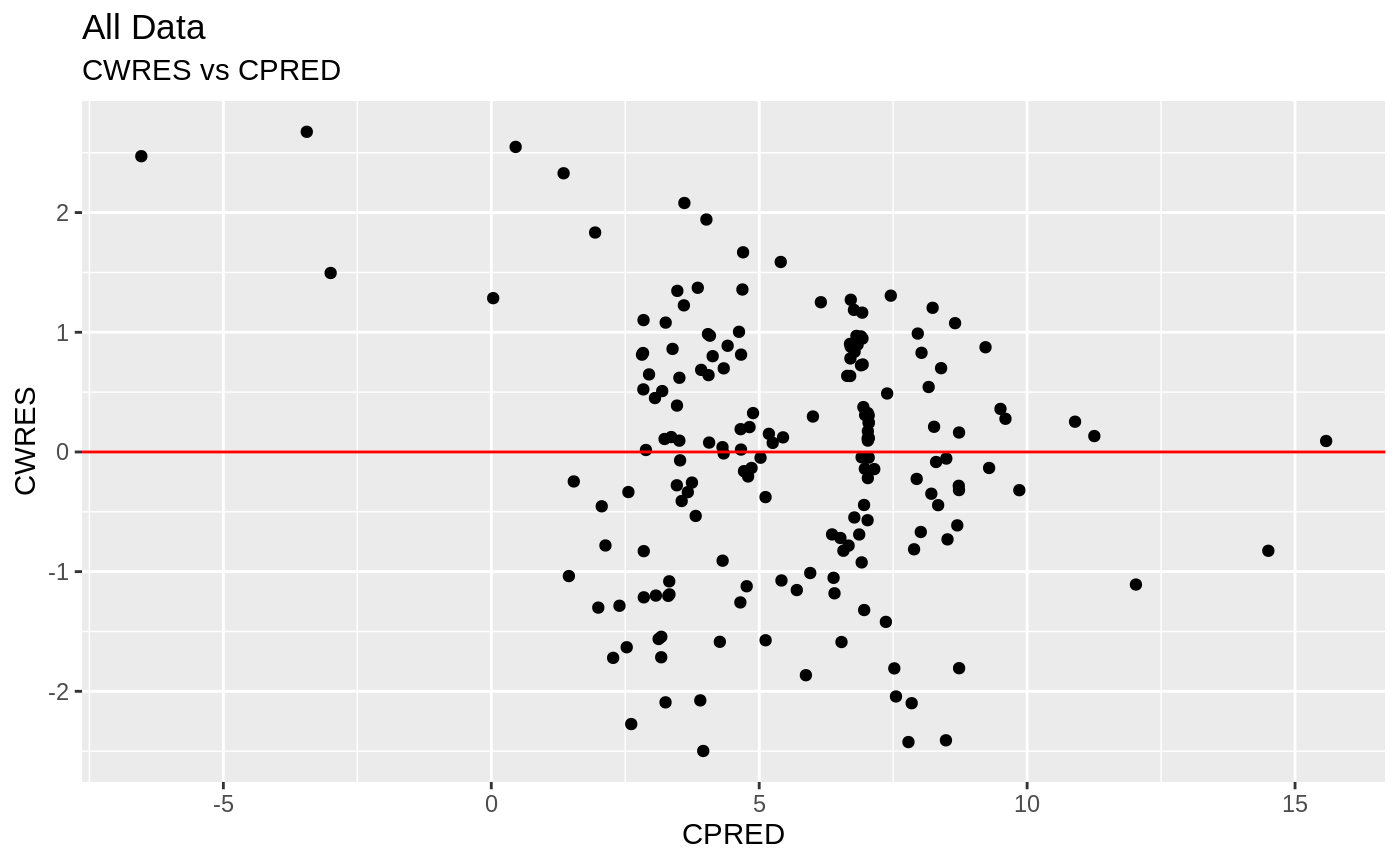
print(res_vs_pred(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))
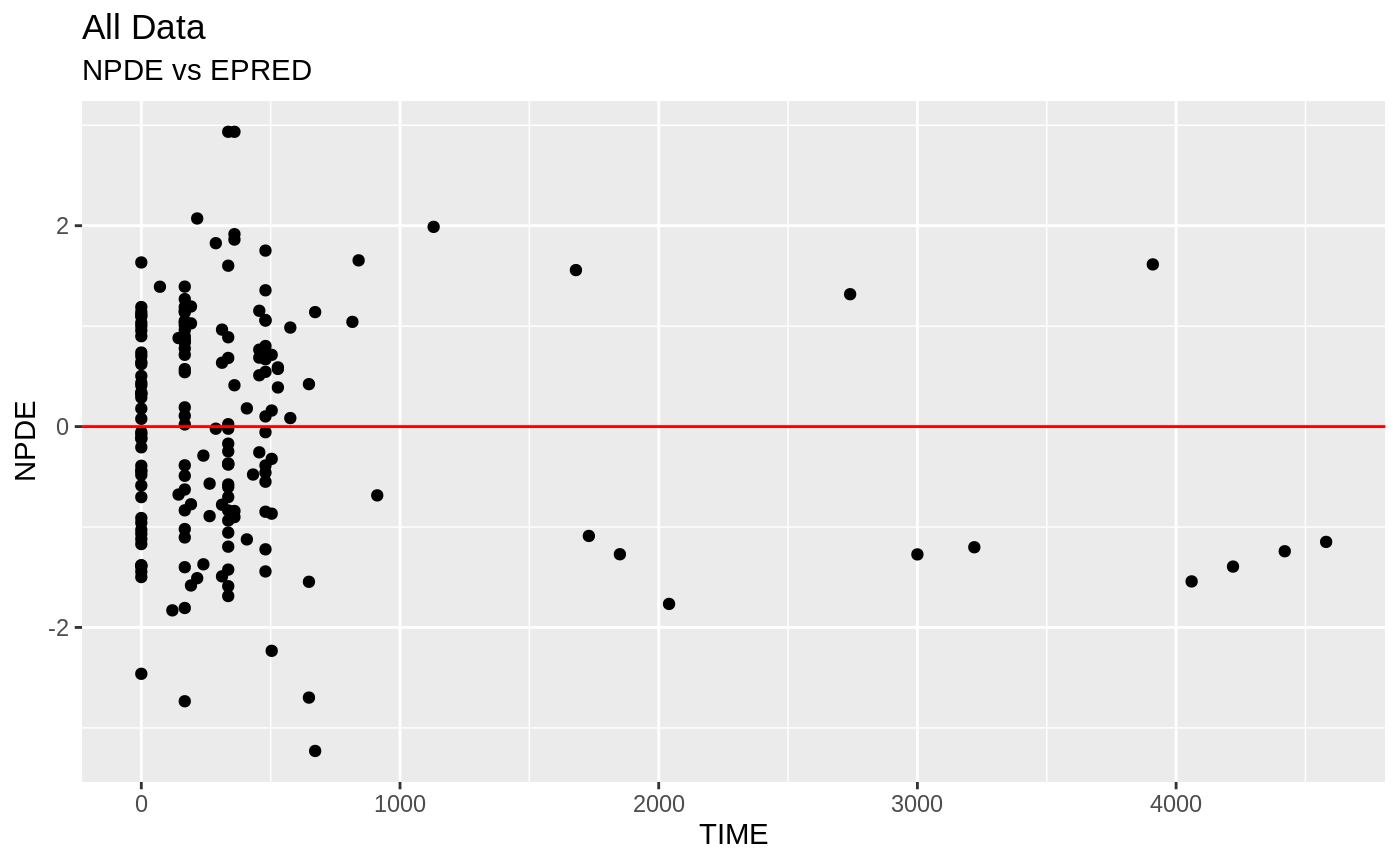
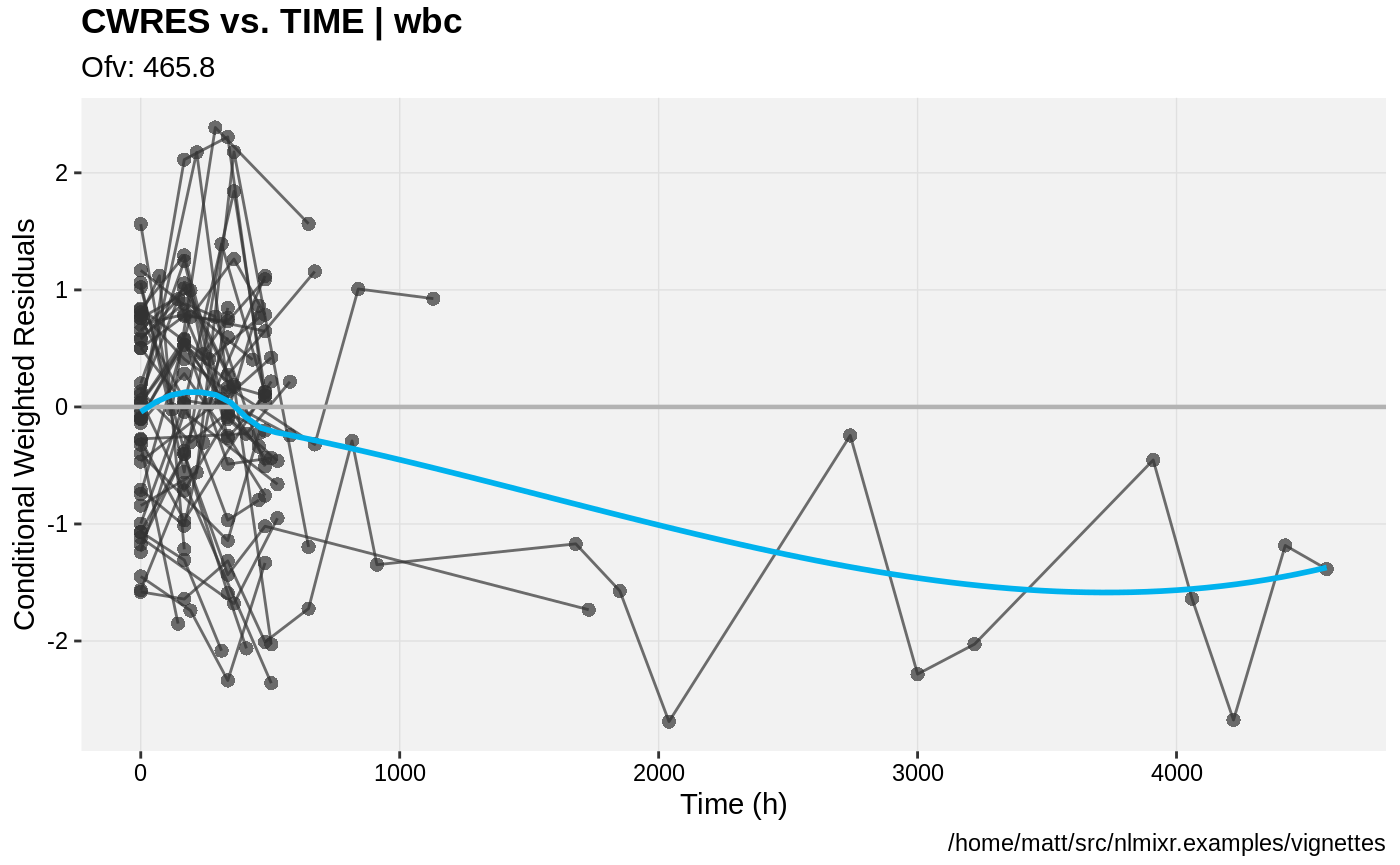
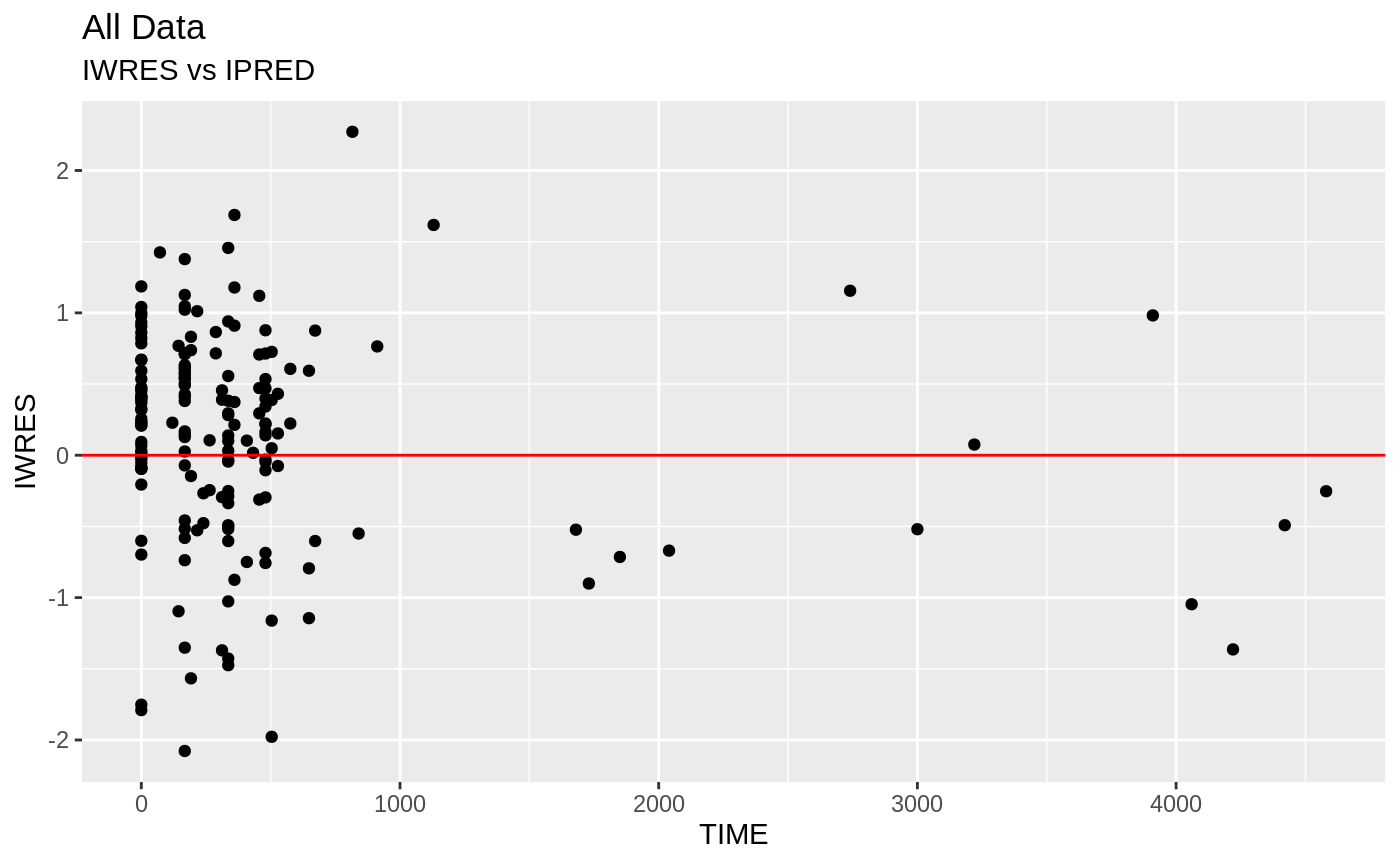
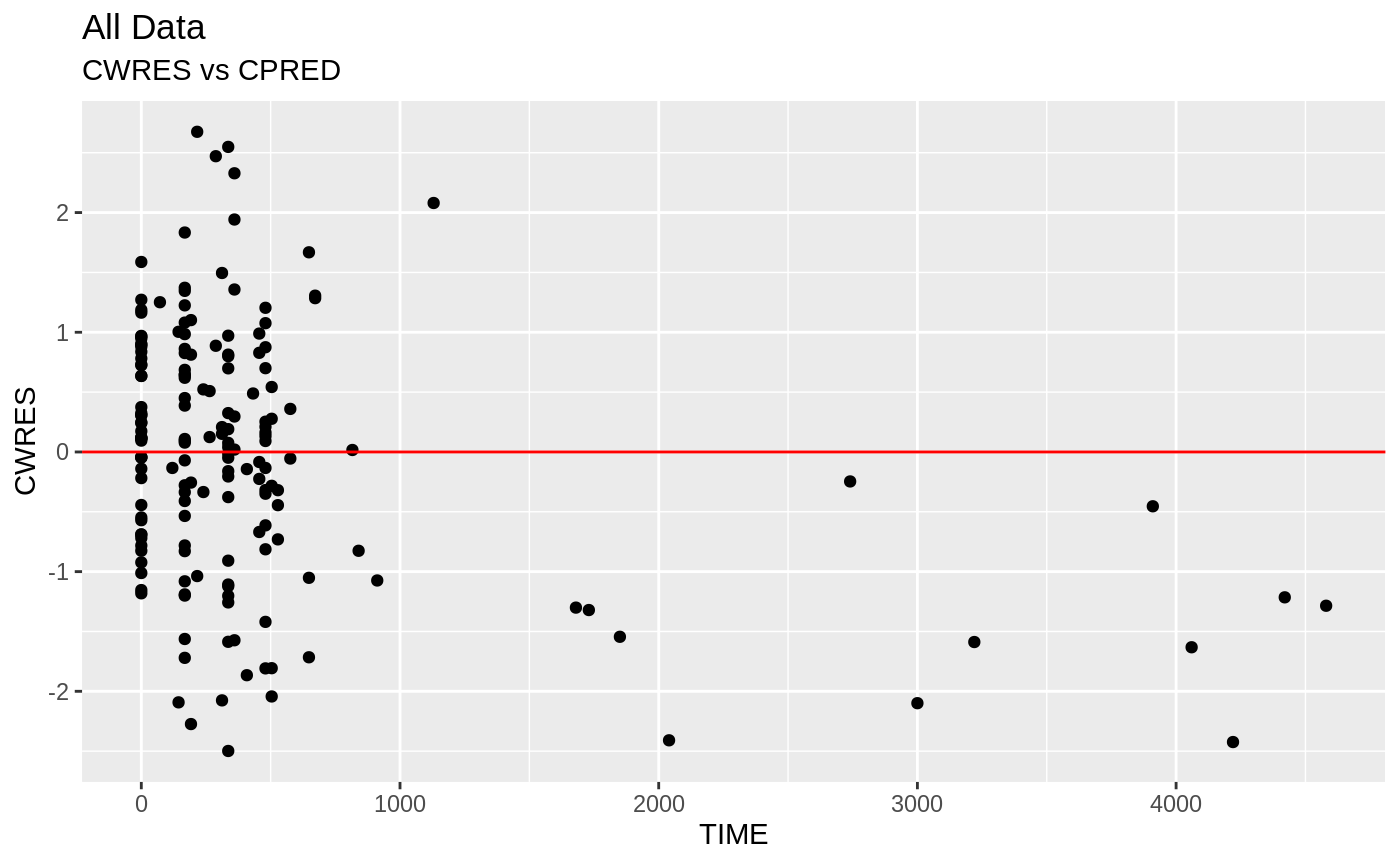
print(res_vs_idv(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Time (h)"))
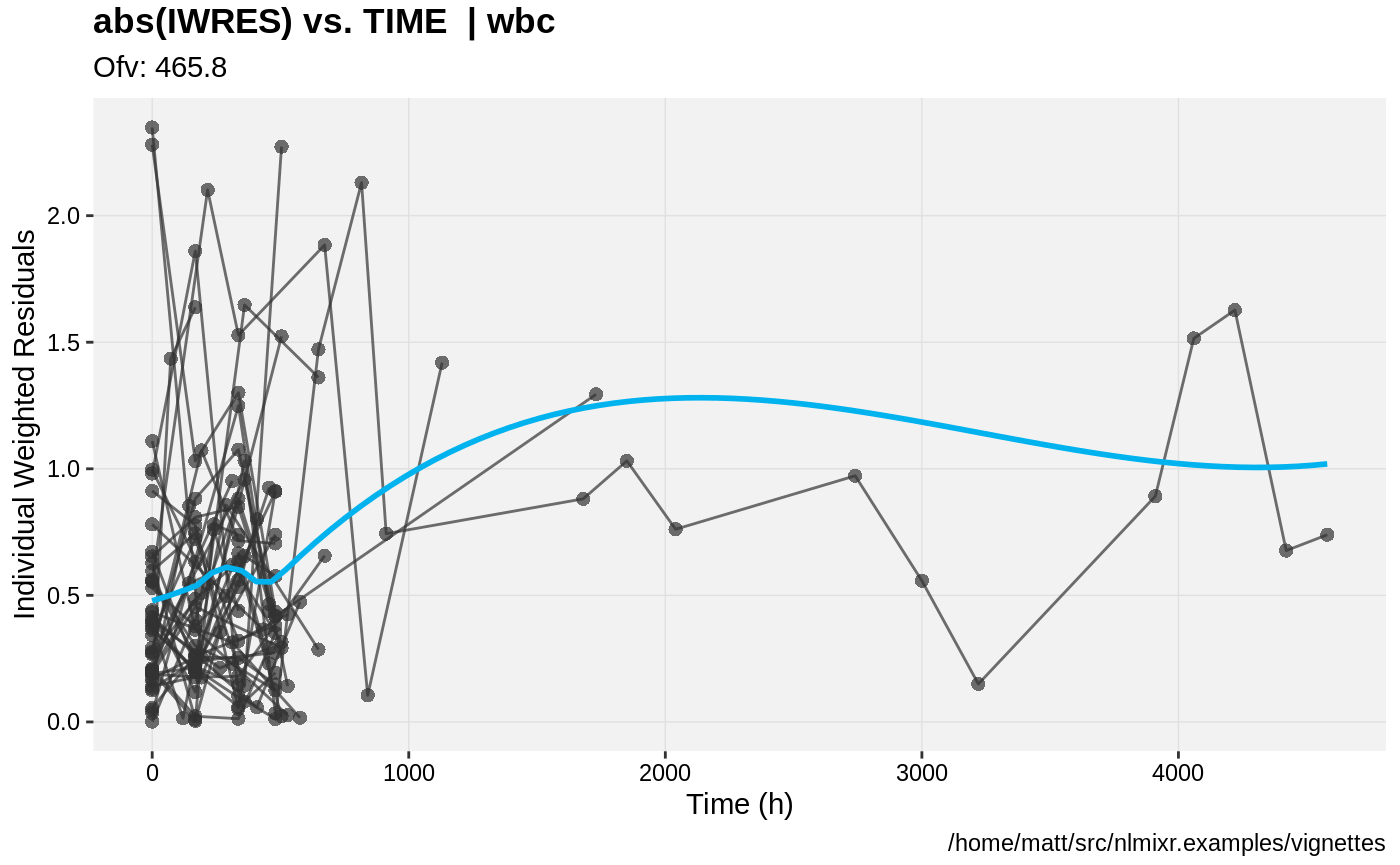
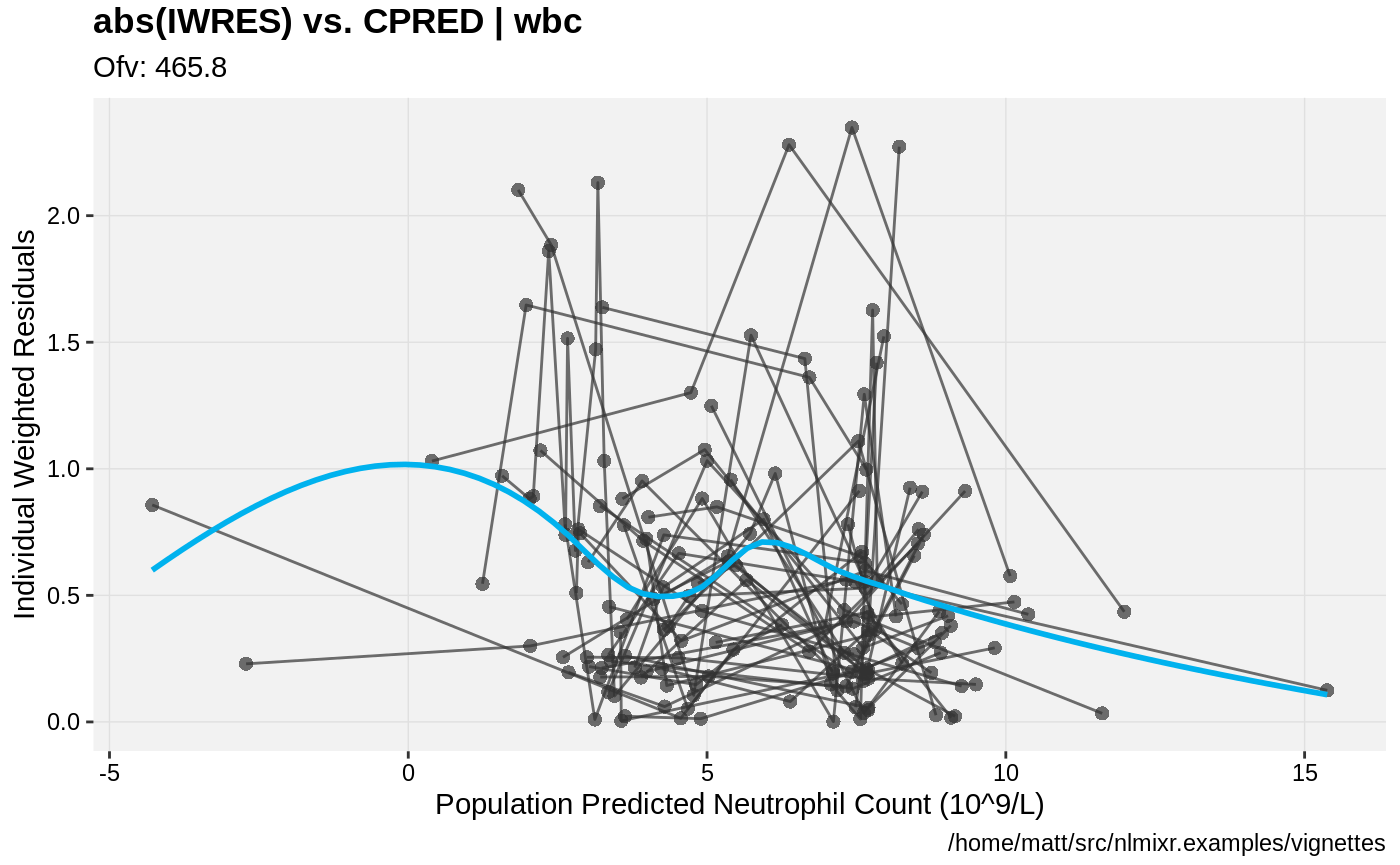
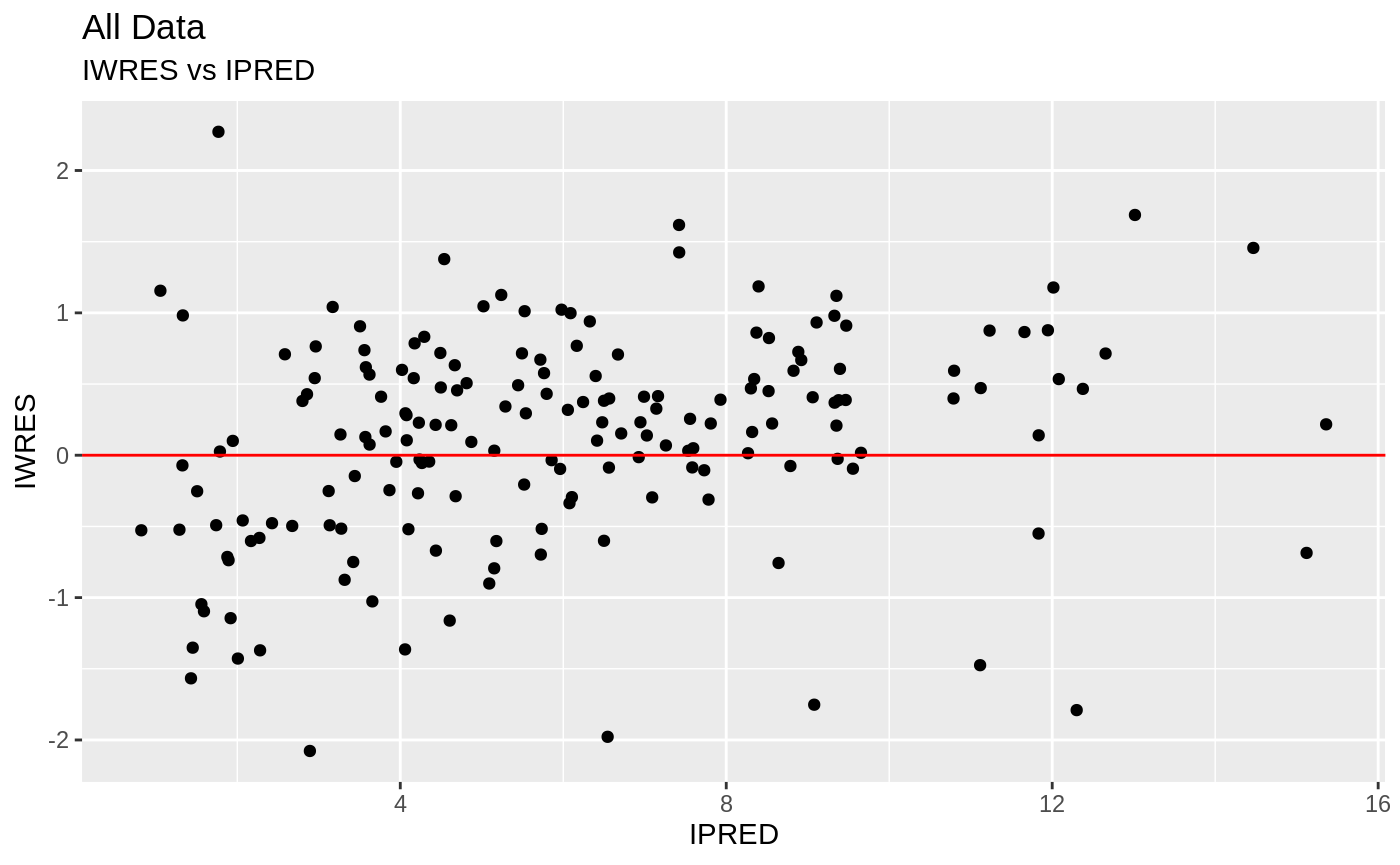
print(absval_res_vs_pred(xpdb, res = 'IWRES') +
ylab("Individual Weighted Residuals") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))
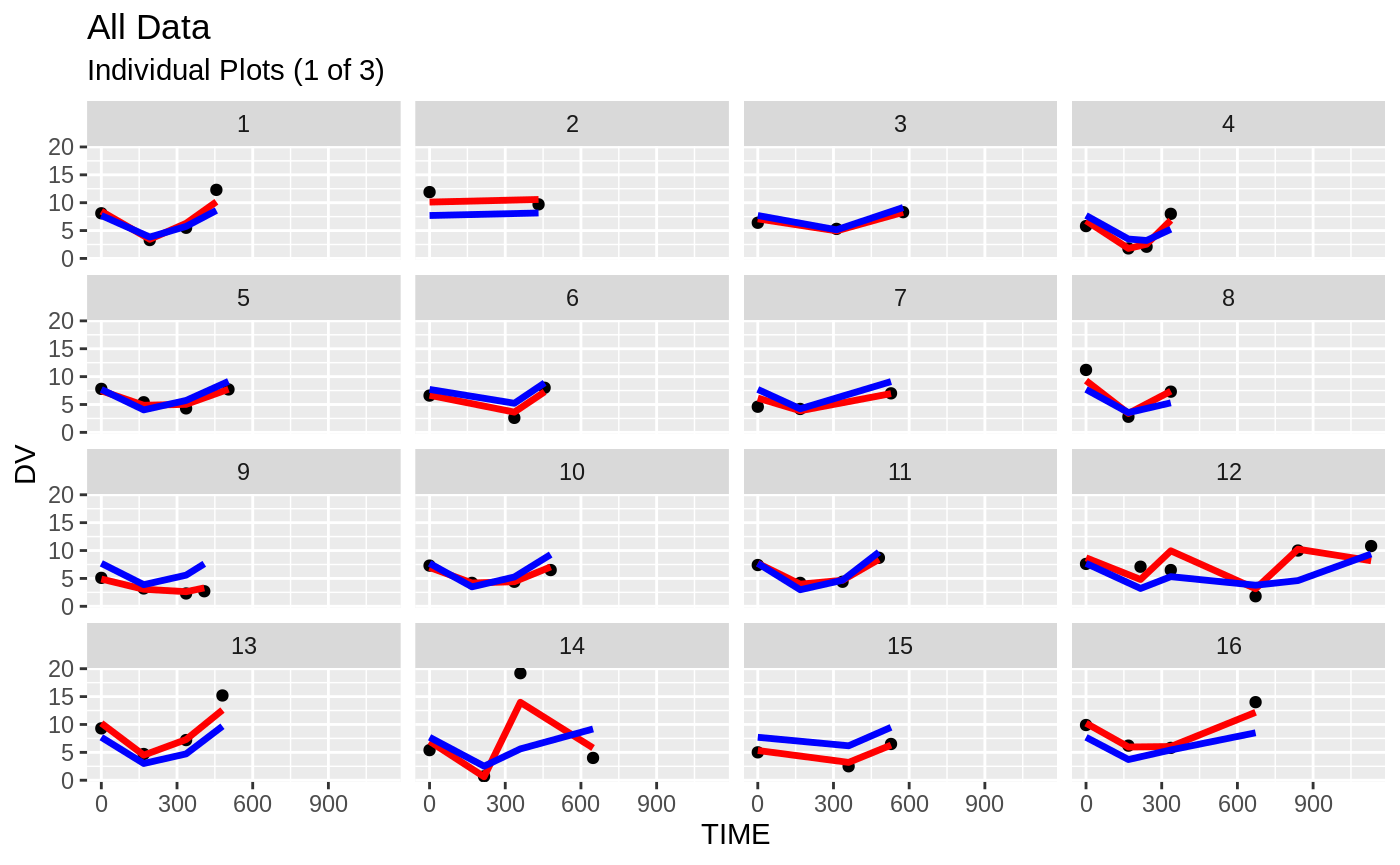
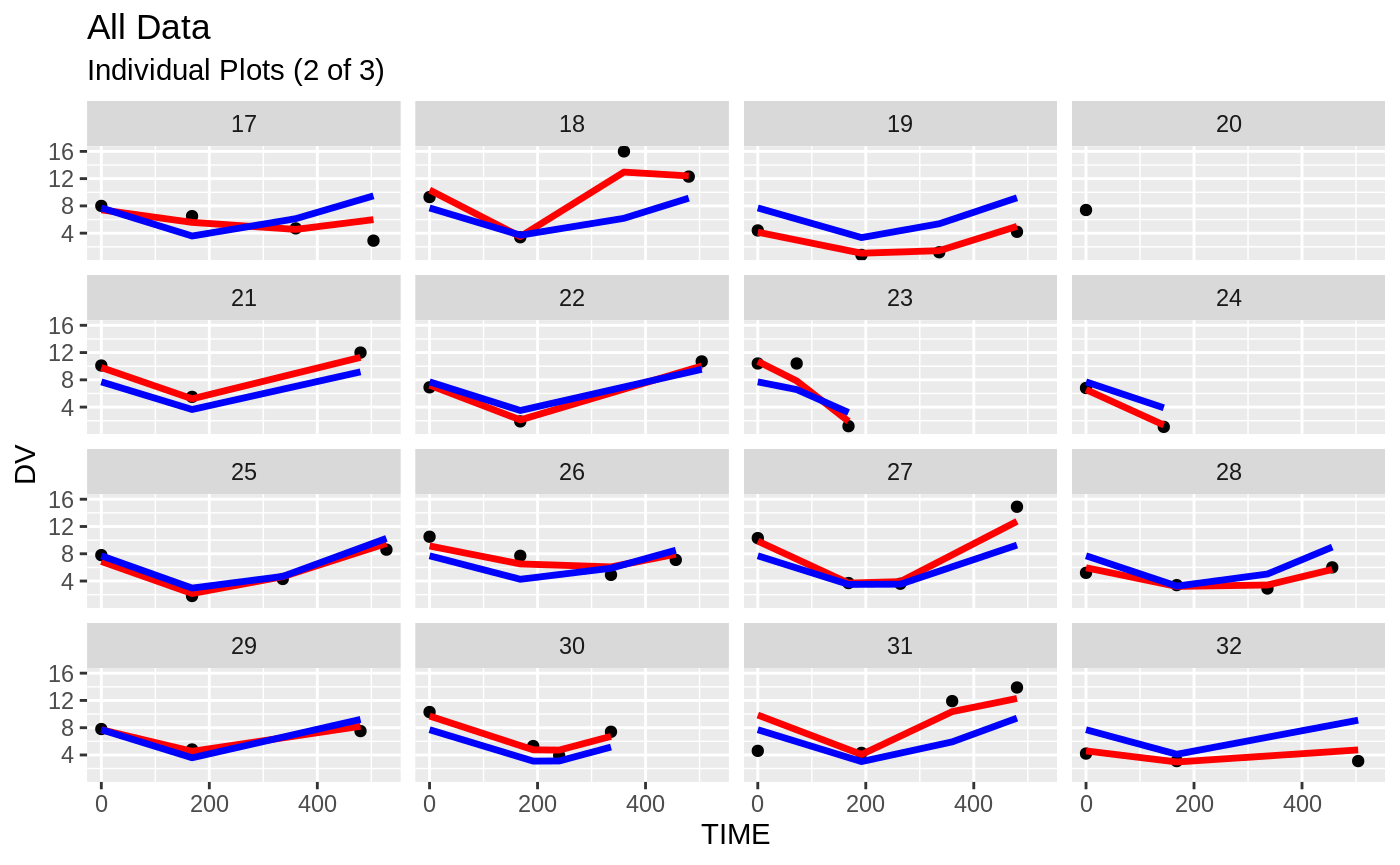
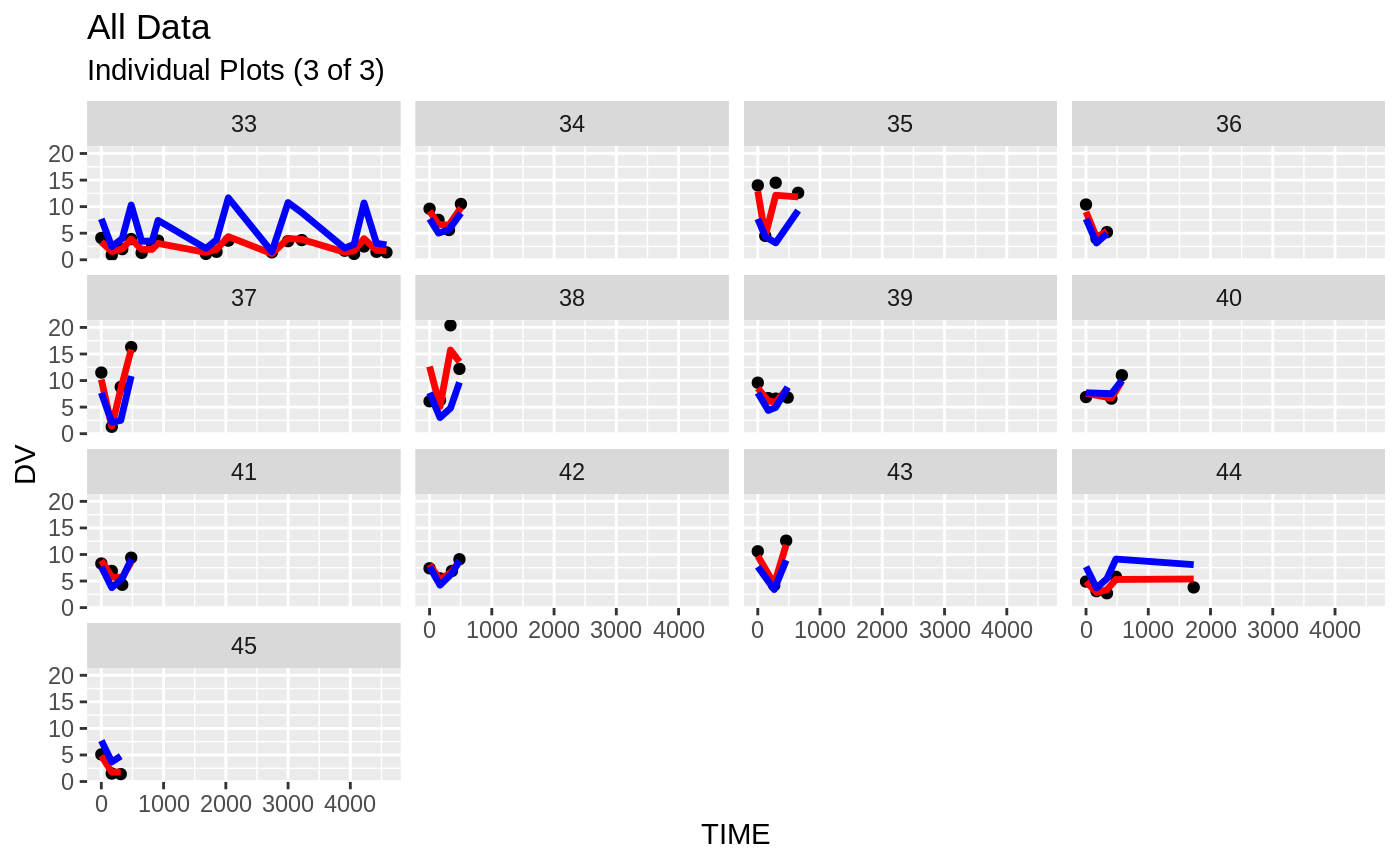
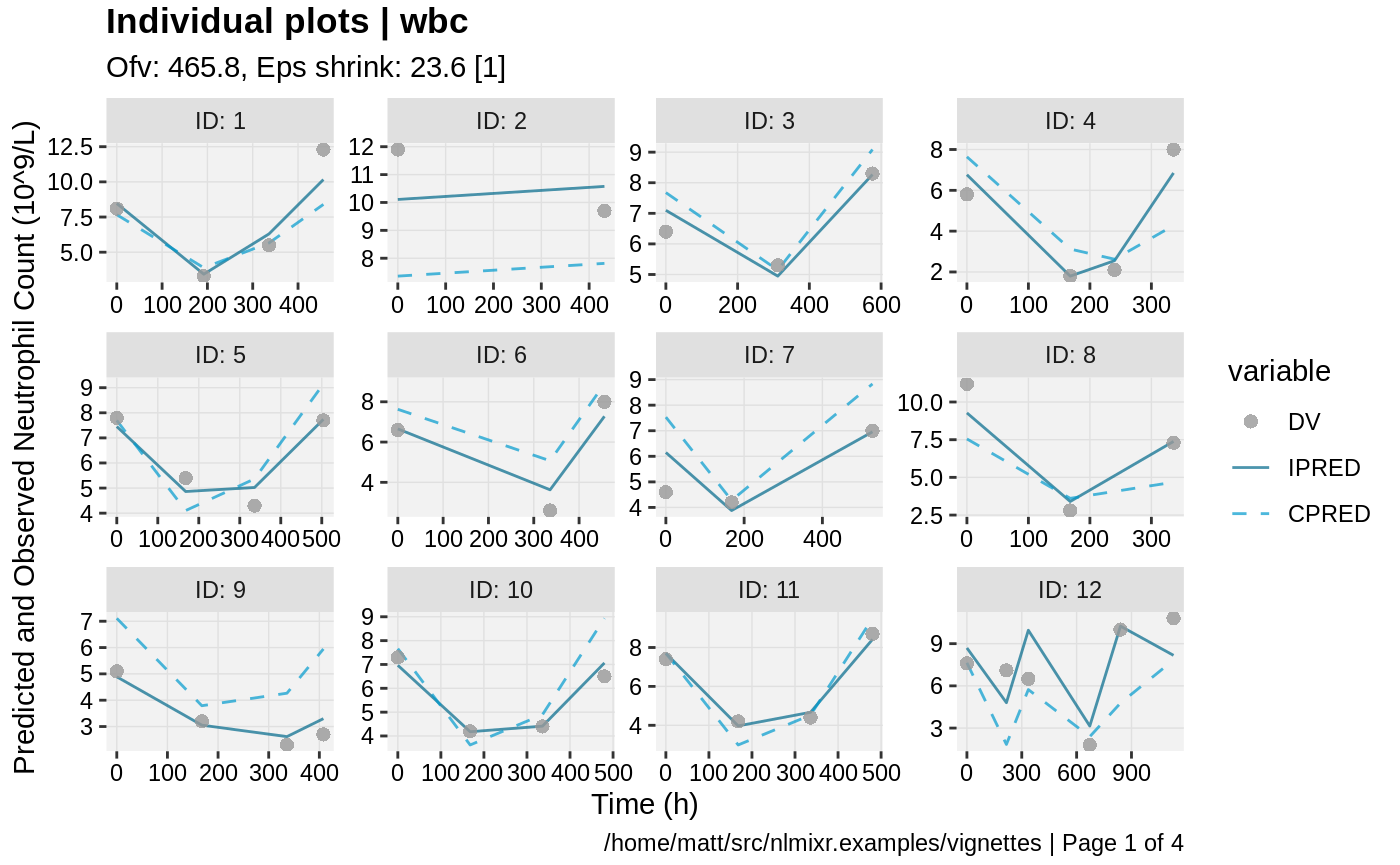
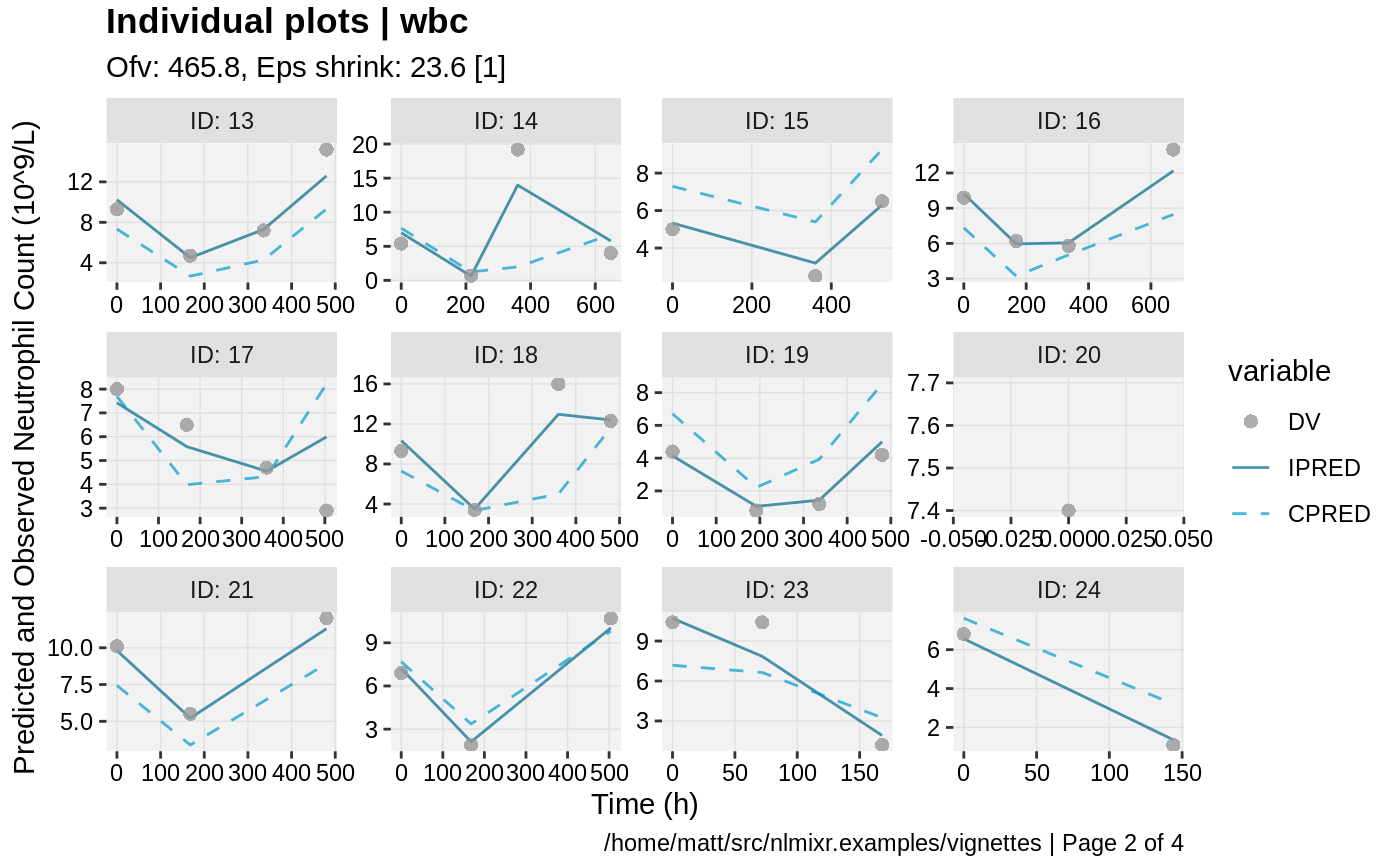
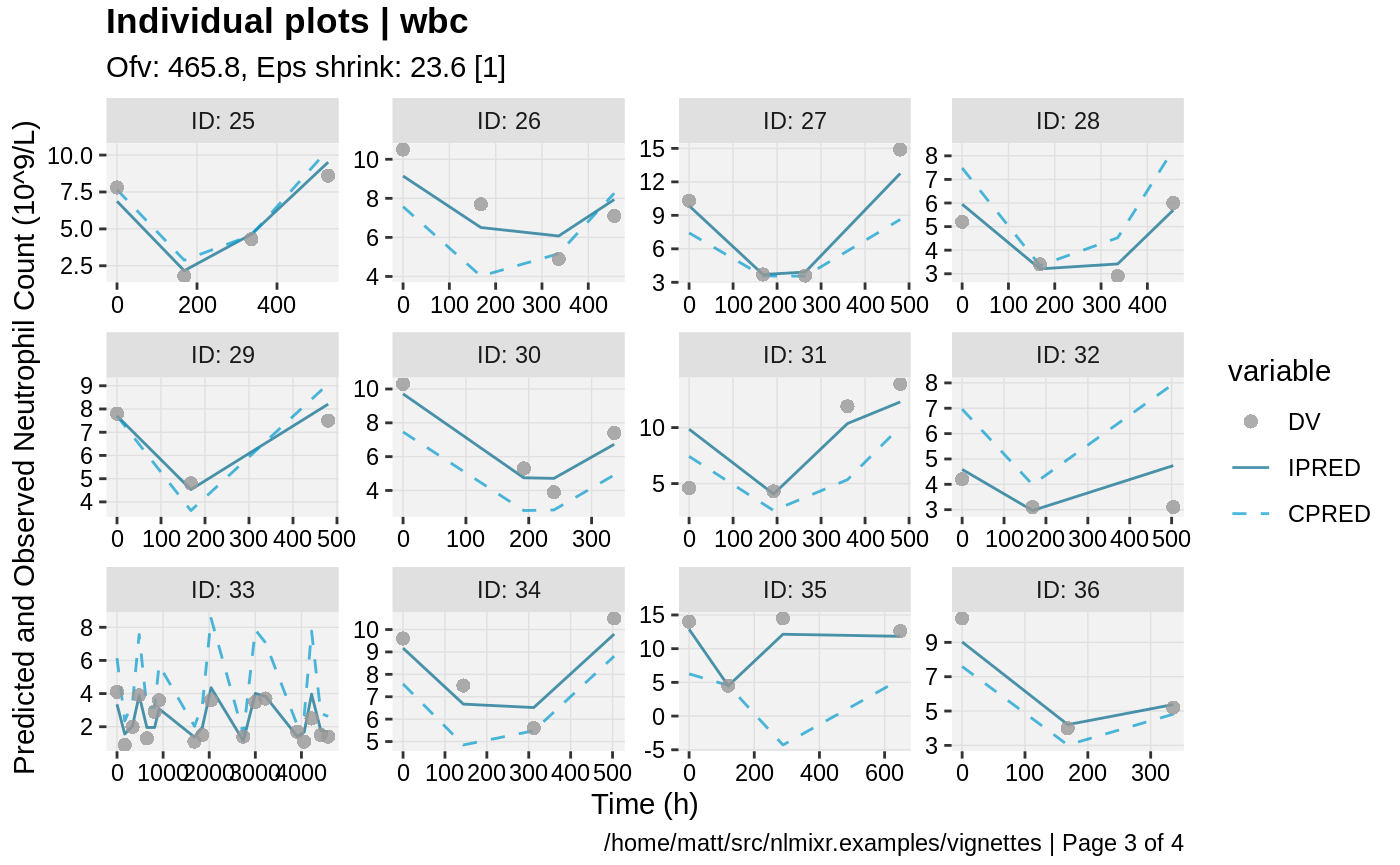
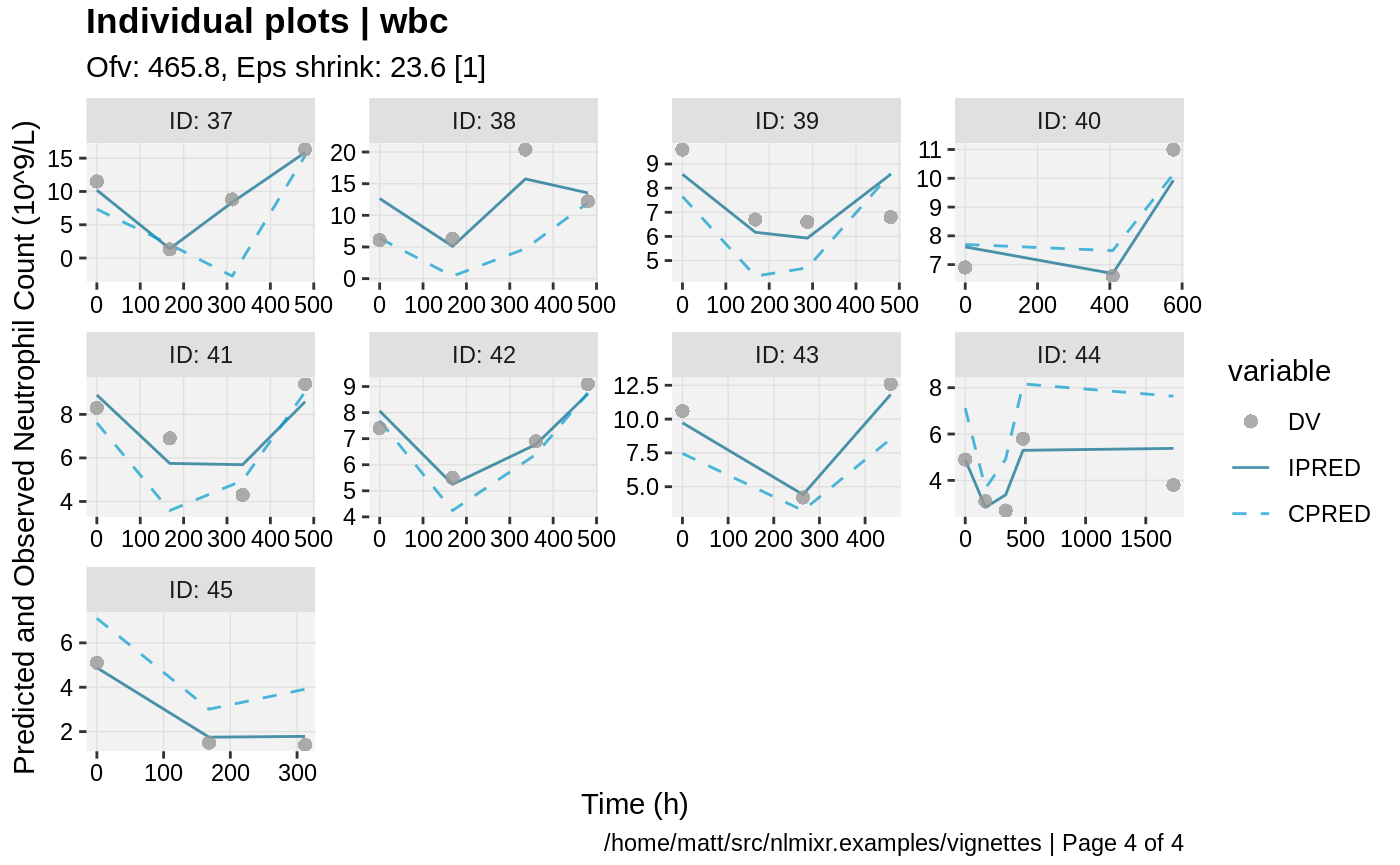
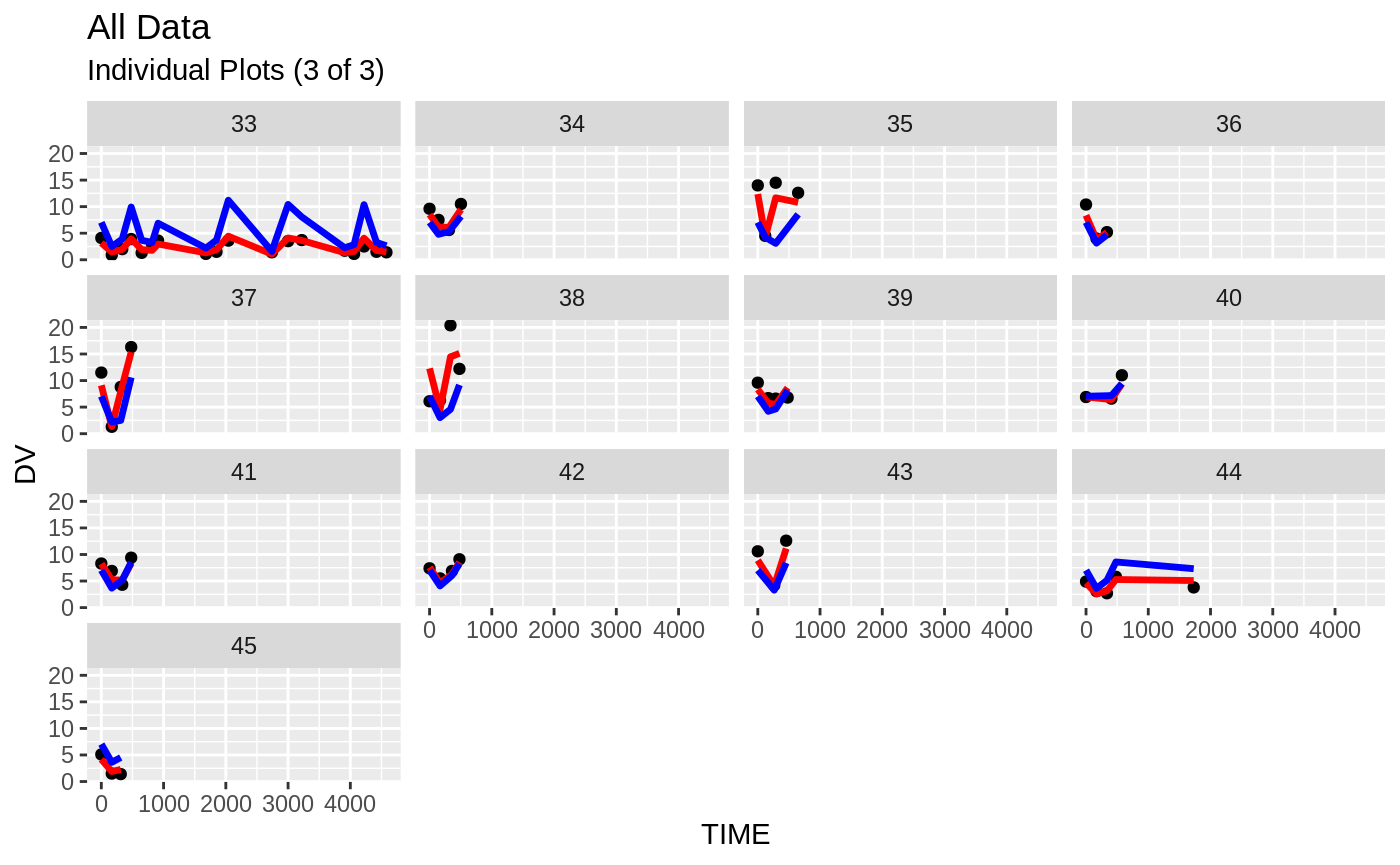
print(ind_plots(xpdb, nrow=3, ncol=4) +
ylab("Predicted and Observed Neutrophil Count (10^9/L)") +
xlab("Time (h)"))
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
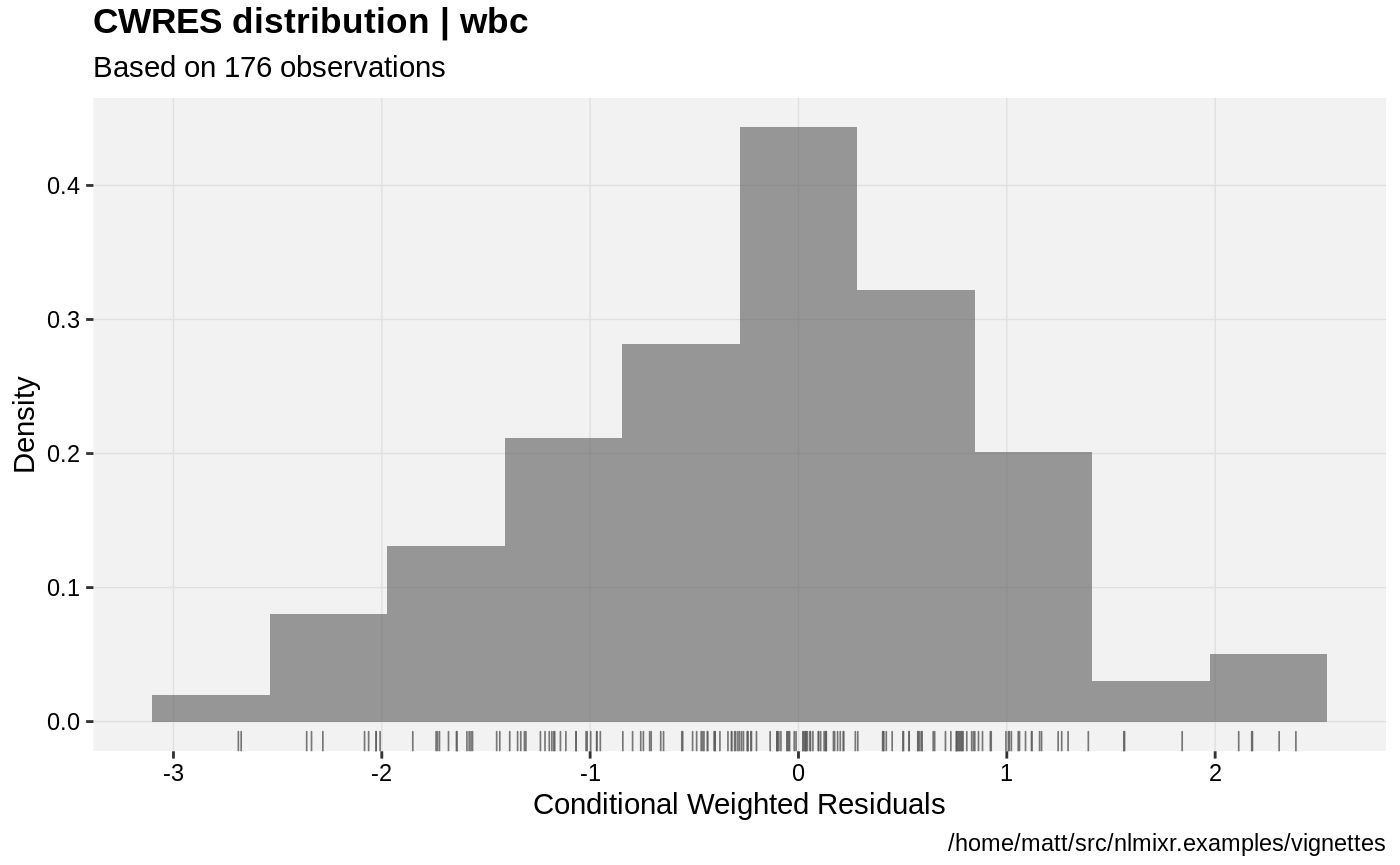
print(res_distrib(xpdb) +
ylab("Density") +
xlab("Conditional Weighted Residuals"))
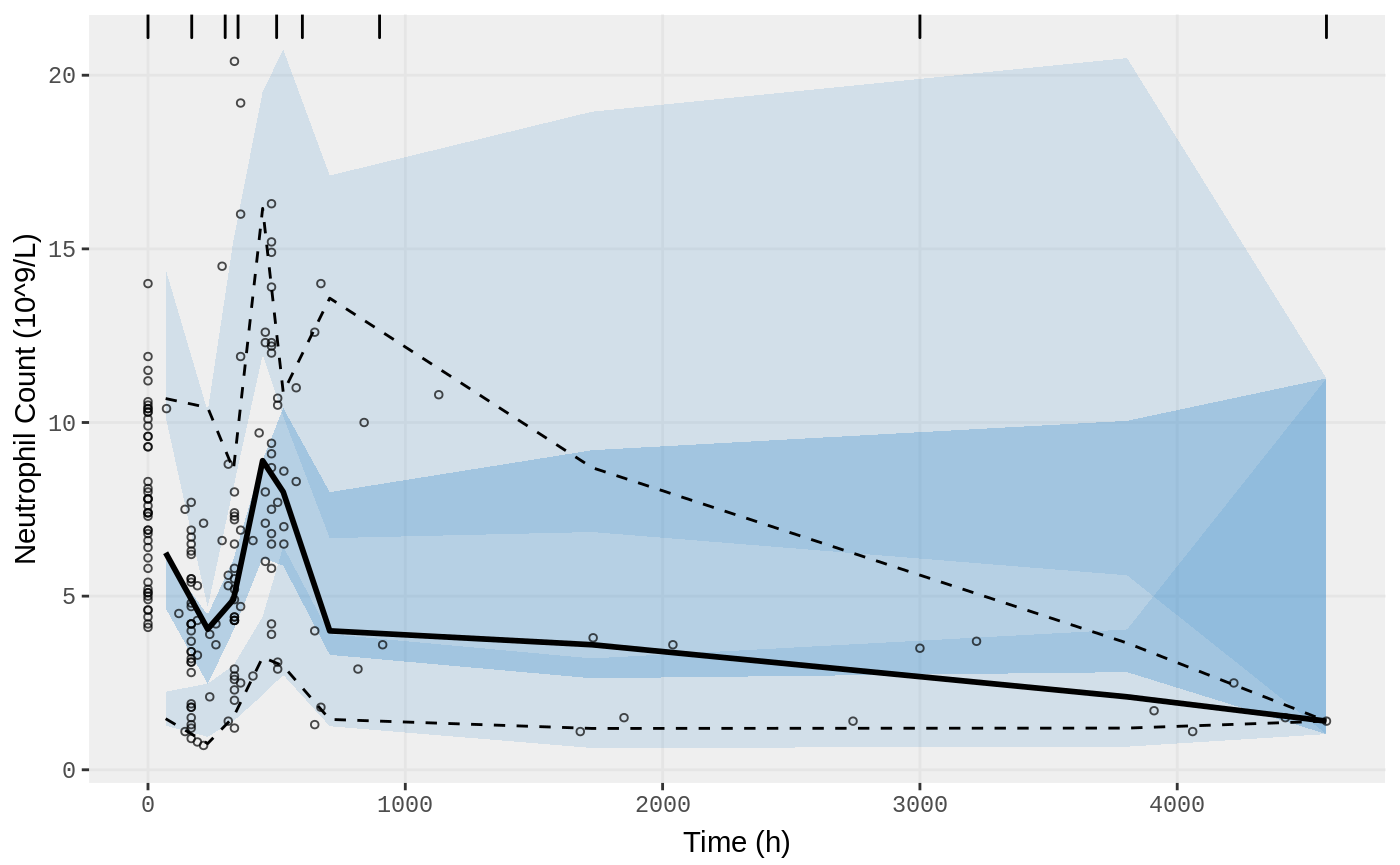
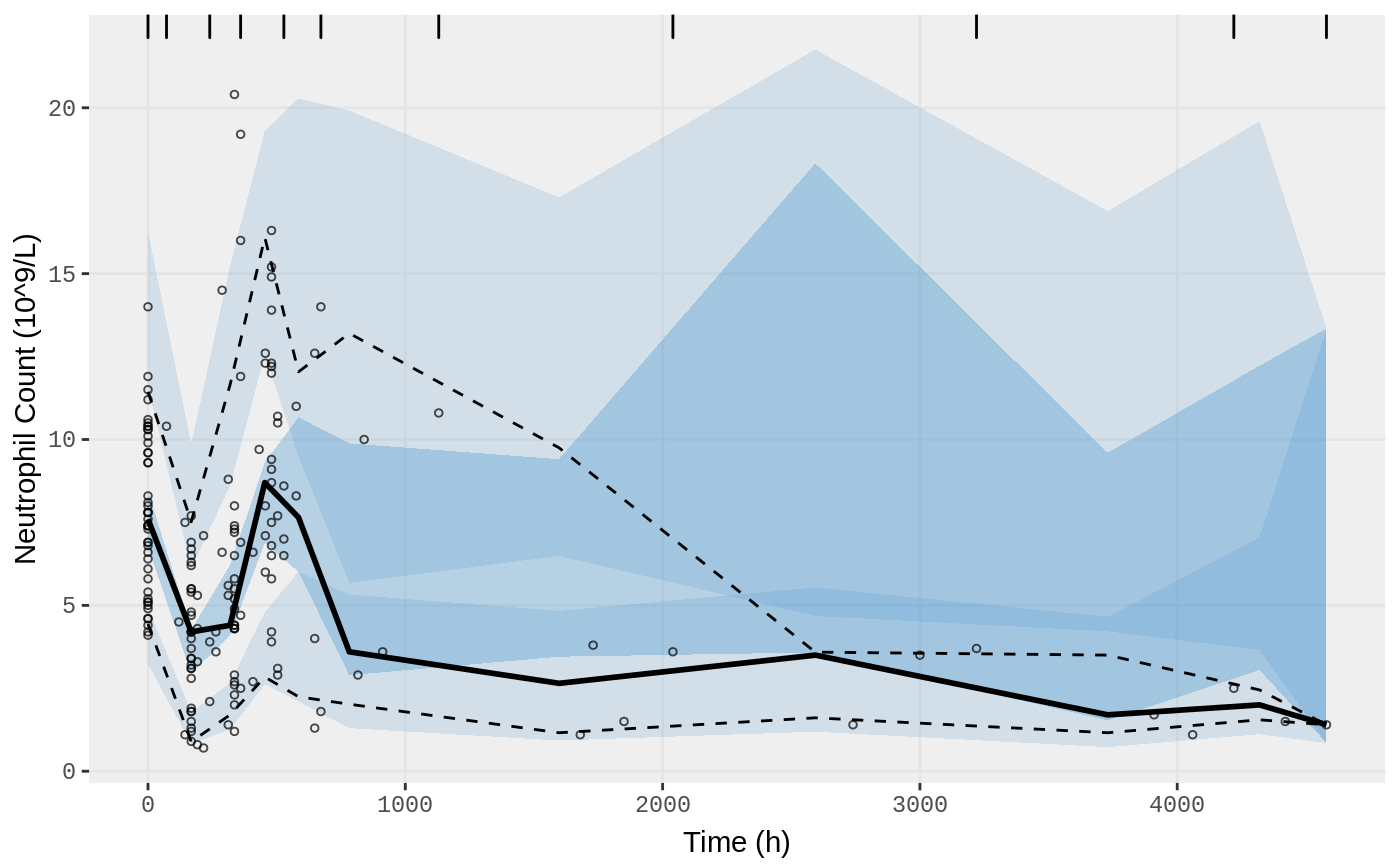
vpc.ui(fit.S, n=500, n_bins = 10, show=list(obs_dv=T), ylab = "Neutrophil Count (10^9/L)", xlab = "Time (h)")
#> Loading vpc already run (/tmp/RtmpXKdUFj/temp_libpath58be457a881a/nlmixr.examples/nlmixr-vpc-wbc-d3--57f2e50dfcdda88e020992f35b58b5f5.rds)
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now
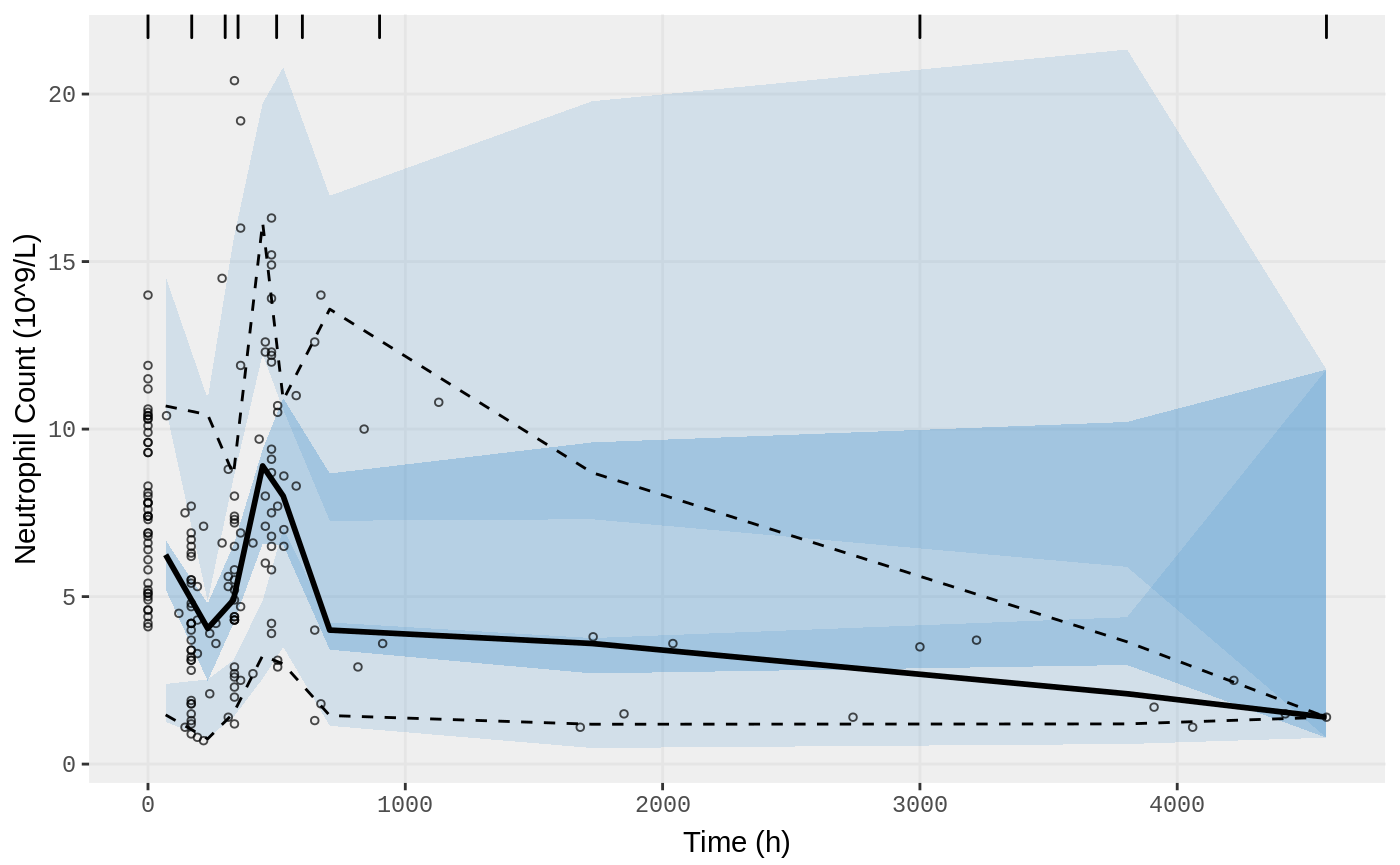
vpc.ui(fit.S, n=500, bins = c(0,170,300,350,500,600,900,3000,4580), show=list(obs_dv=T), ylab = "Neutrophil Count (10^9/L)", xlab = "Time (h)")
#> Loading vpc already run (/tmp/RtmpXKdUFj/temp_libpath58be457a881a/nlmixr.examples/nlmixr-vpc-wbc-d3--cabedf9123bd16bc37f8f5f419231637.rds)
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now
Fit model using FOCEi
fit.F <- nlmixr(wbc, d3, est="focei", table=list(cwres=TRUE, npde=TRUE))
#> Loading model already run (/tmp/RtmpXKdUFj/temp_libpath58be457a881a/nlmixr.examples/nlmixr-wbc-d3-focei-36419c3f3c6bcd0192bae24b06db4138.rds)
#> Warning in lapply(X = X, FUN = FUN, ...): ID missing in parameters dataset;
#> Parameters are assumed to have the same order as the IDs in the event dataset
#> Warning in lapply(X = X, FUN = FUN, ...): Initial ETAs were nudged; (Can
#> control by foceiControl(etaNudge=.))
#> Warning in lapply(X = X, FUN = FUN, ...): ETAs were reset to zero during
#> optimization; (Can control by foceiControl(resetEtaP=.))
#> Warning in lapply(X = X, FUN = FUN, ...): Gradient problems with initial
#> estimate and covariance; see $scaleInfo
#> Warning in lapply(X = X, FUN = FUN, ...): Tolerances (atol/rtol) were reduced (after 5 bad solves) for some difficult ODE solving during the optimization.
#> Can control with foceiControl(stickyRecalcN=)
#> Consider reducing sigdig/atol/rtol changing initial estimates or changing the structural model.
FOCEi Goodness of fit plots
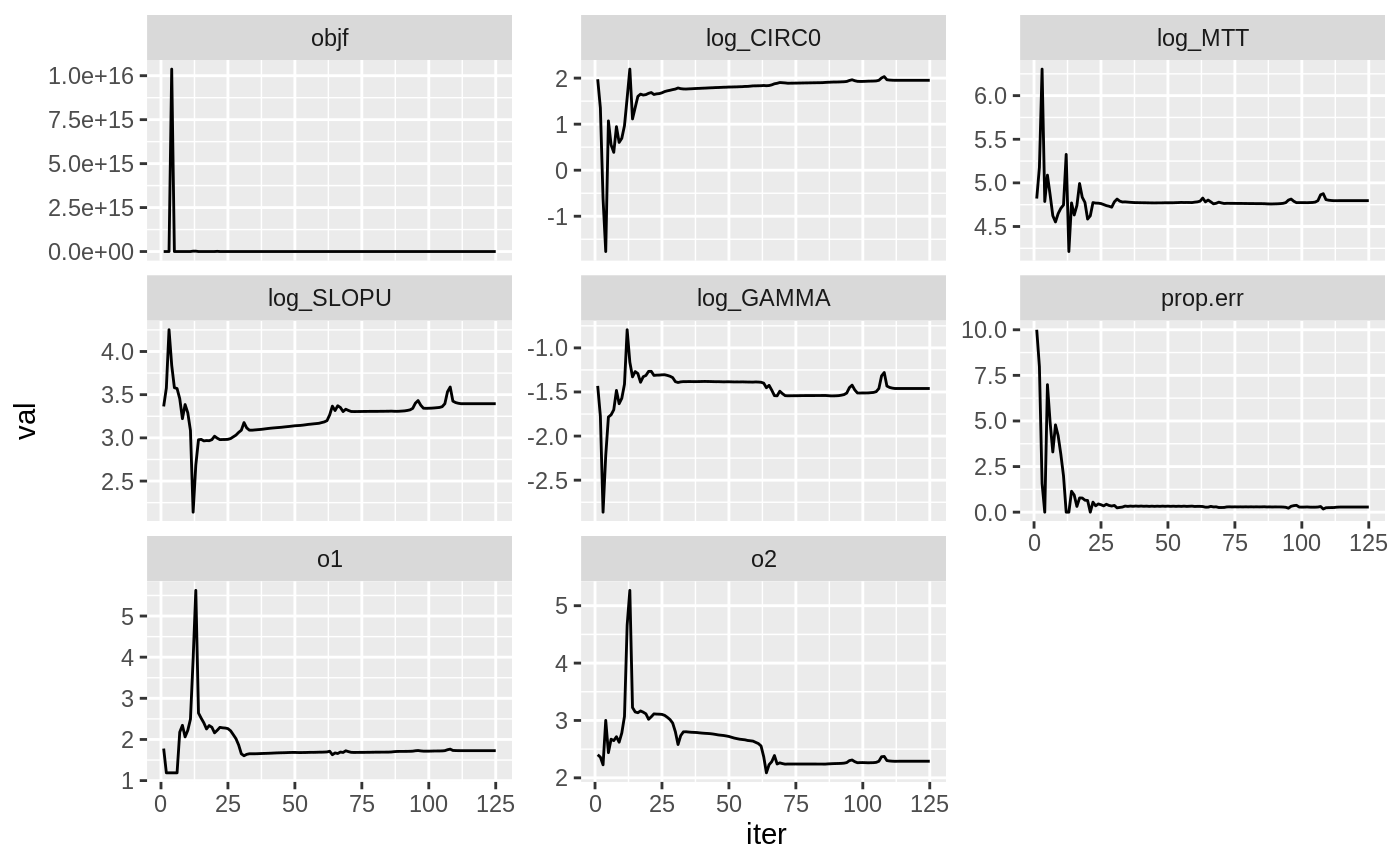
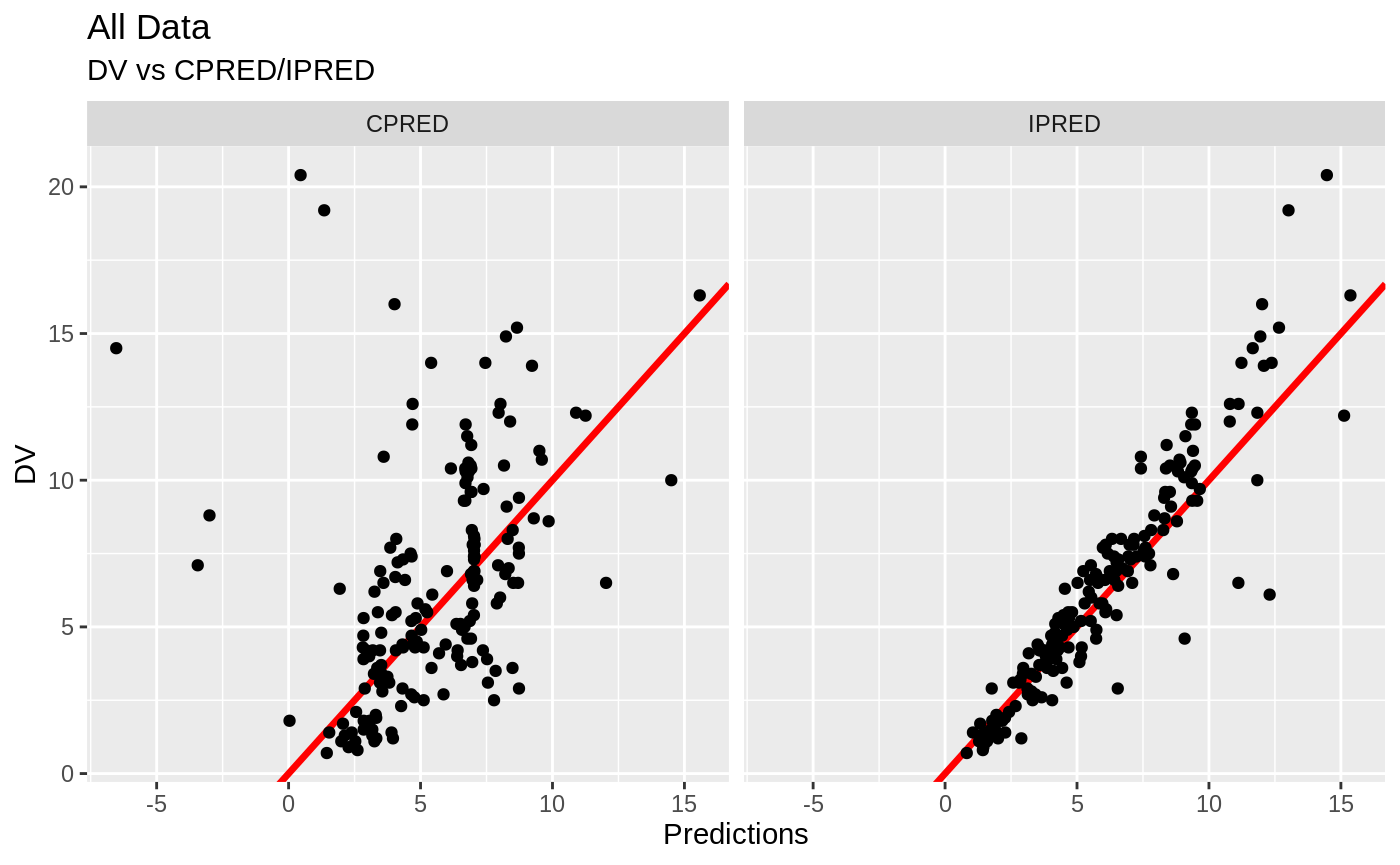
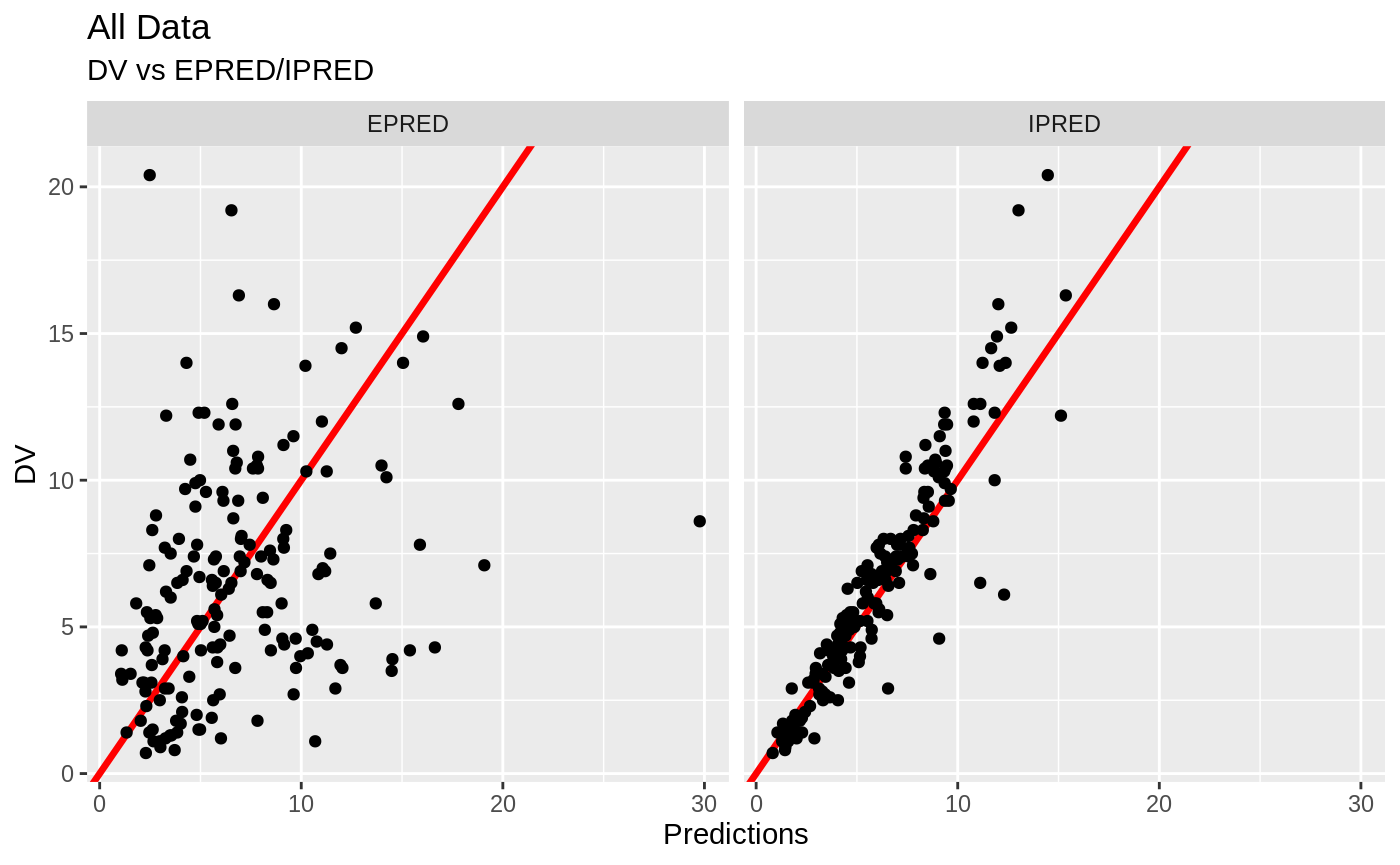
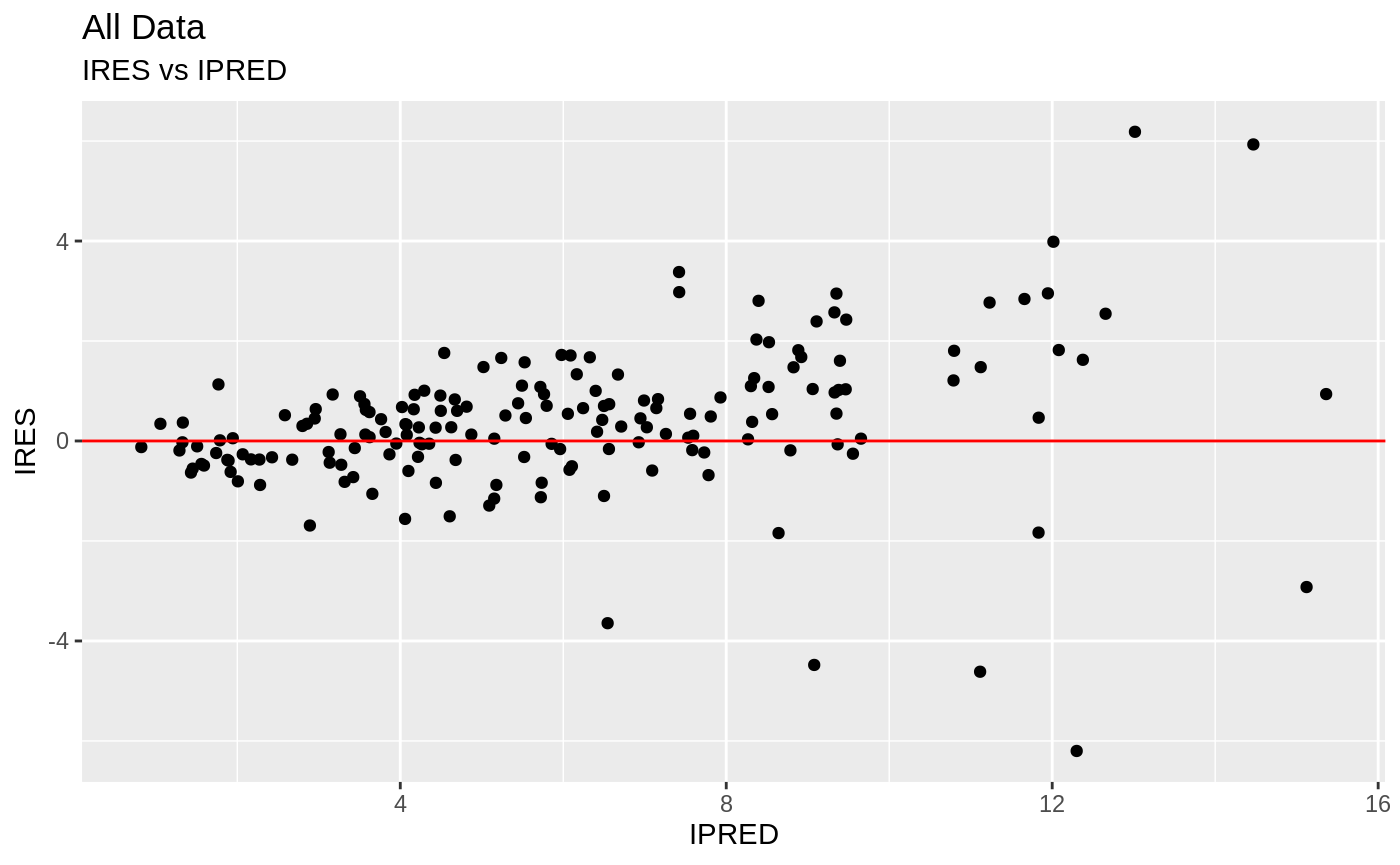
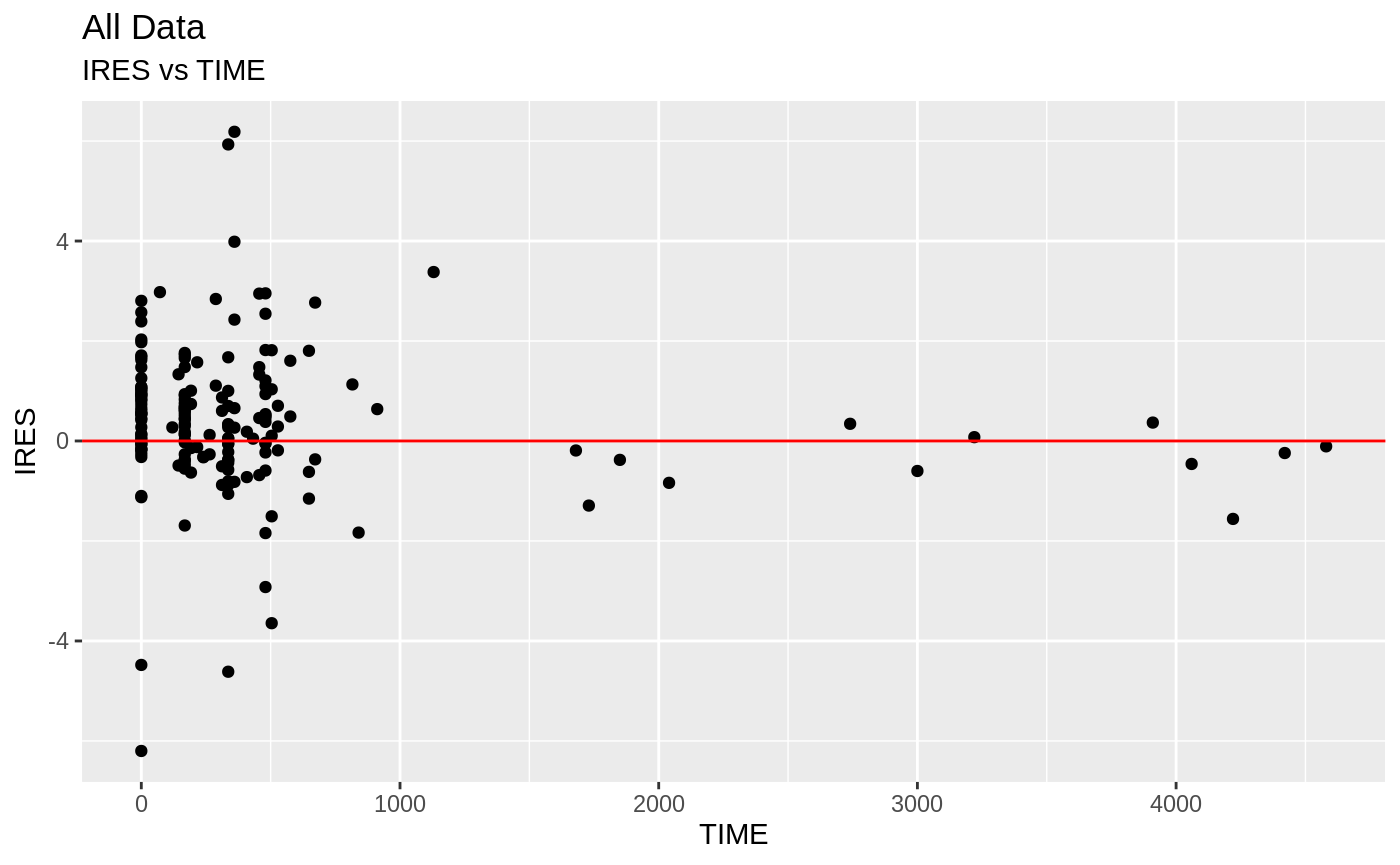
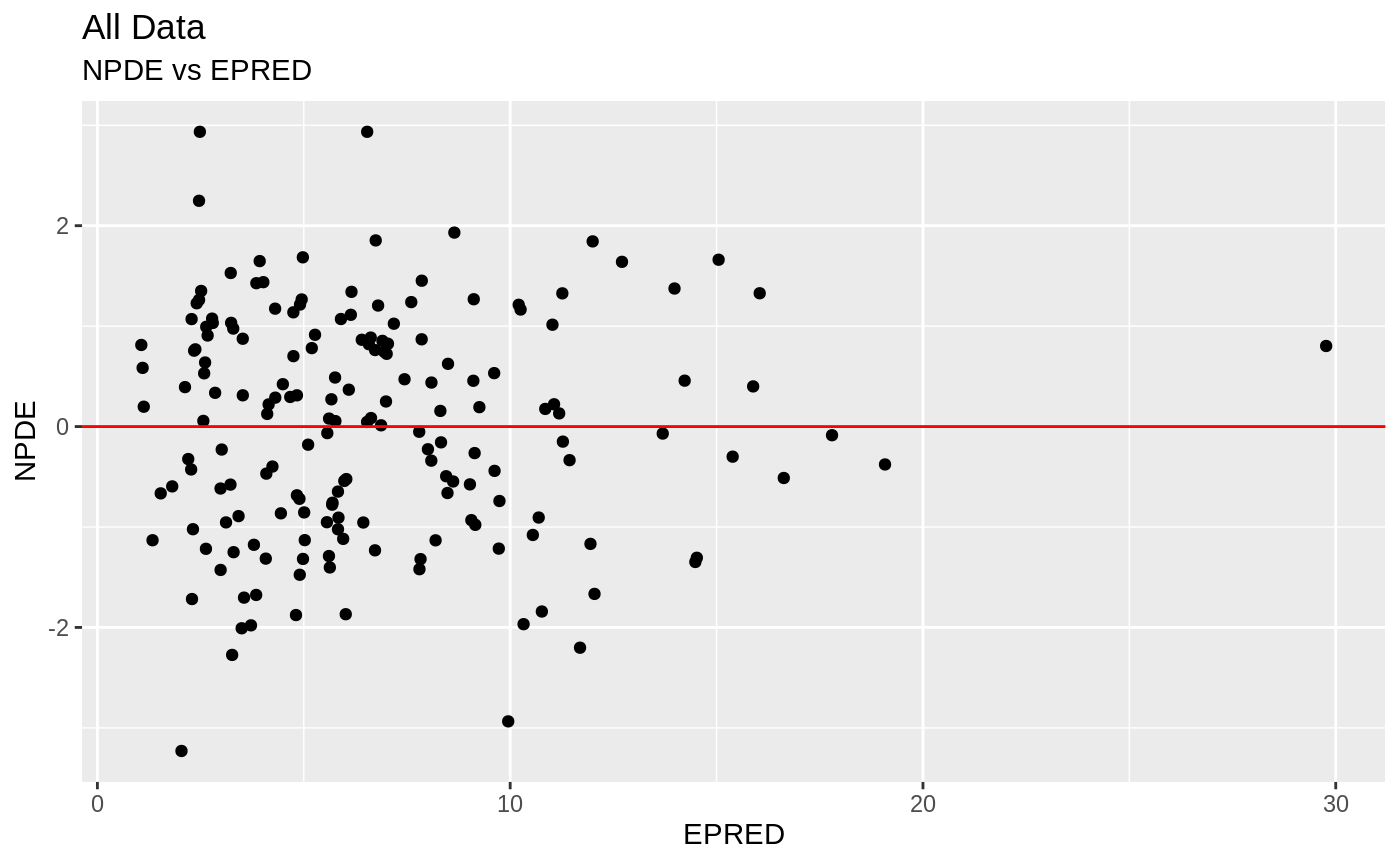
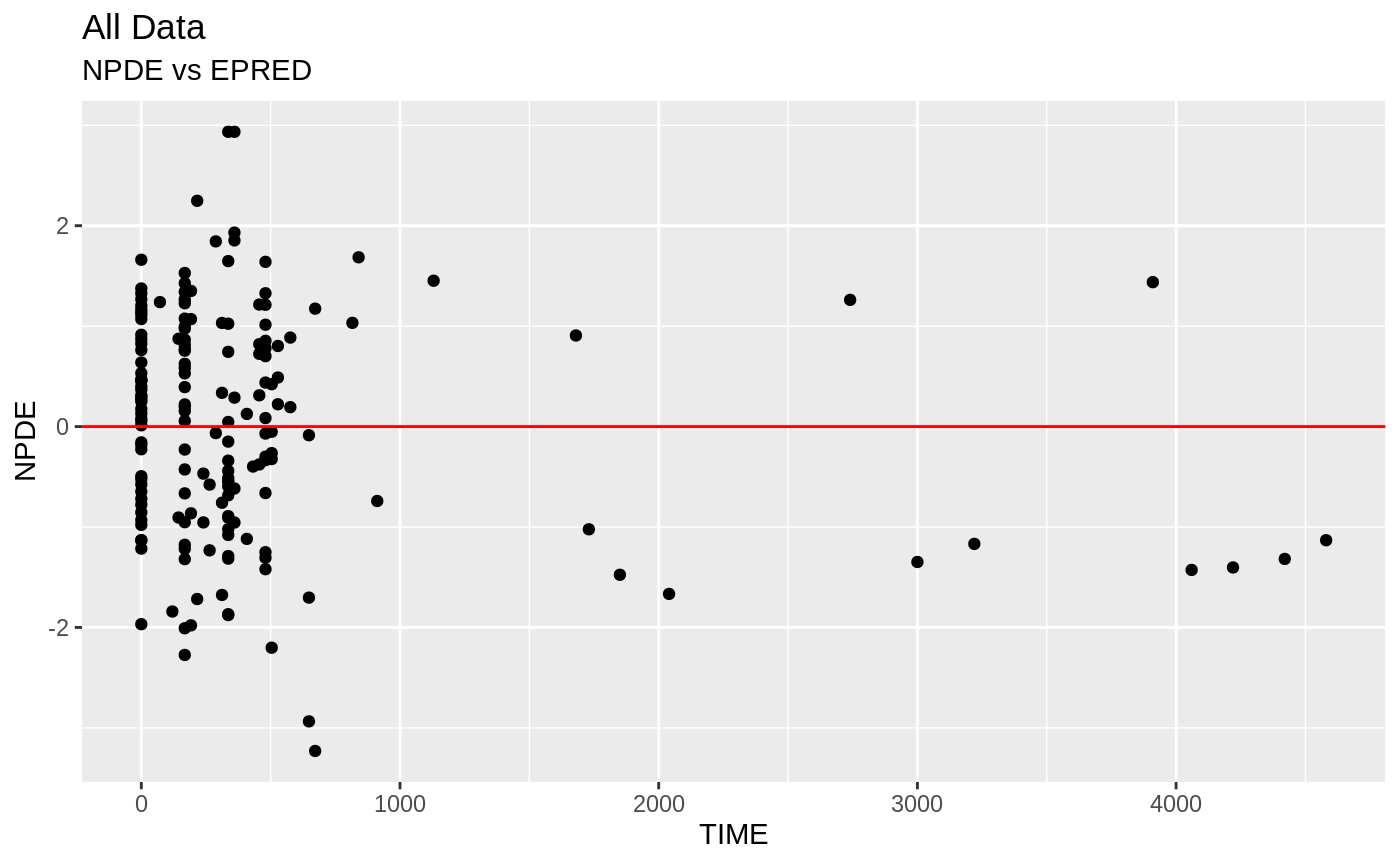
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
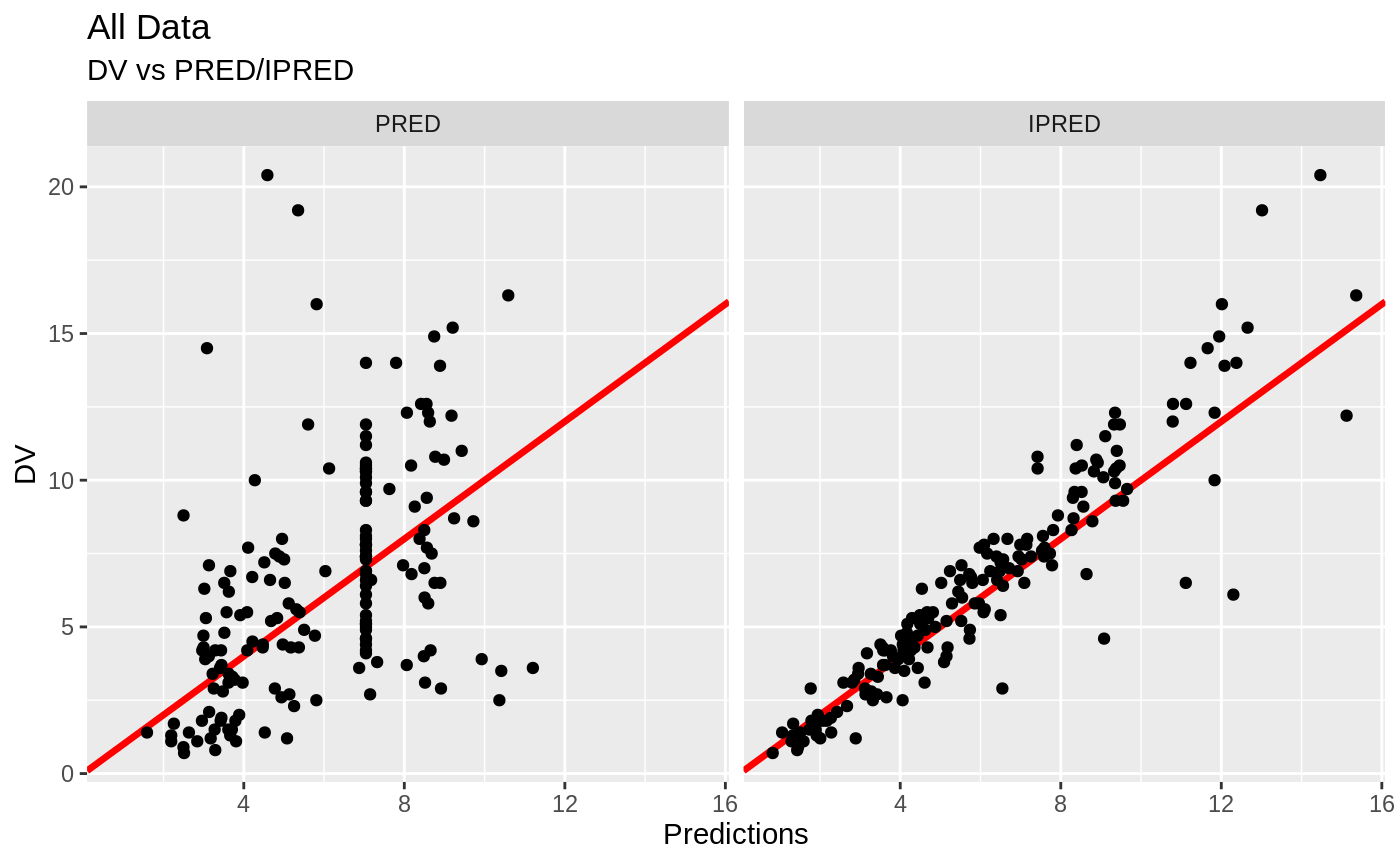
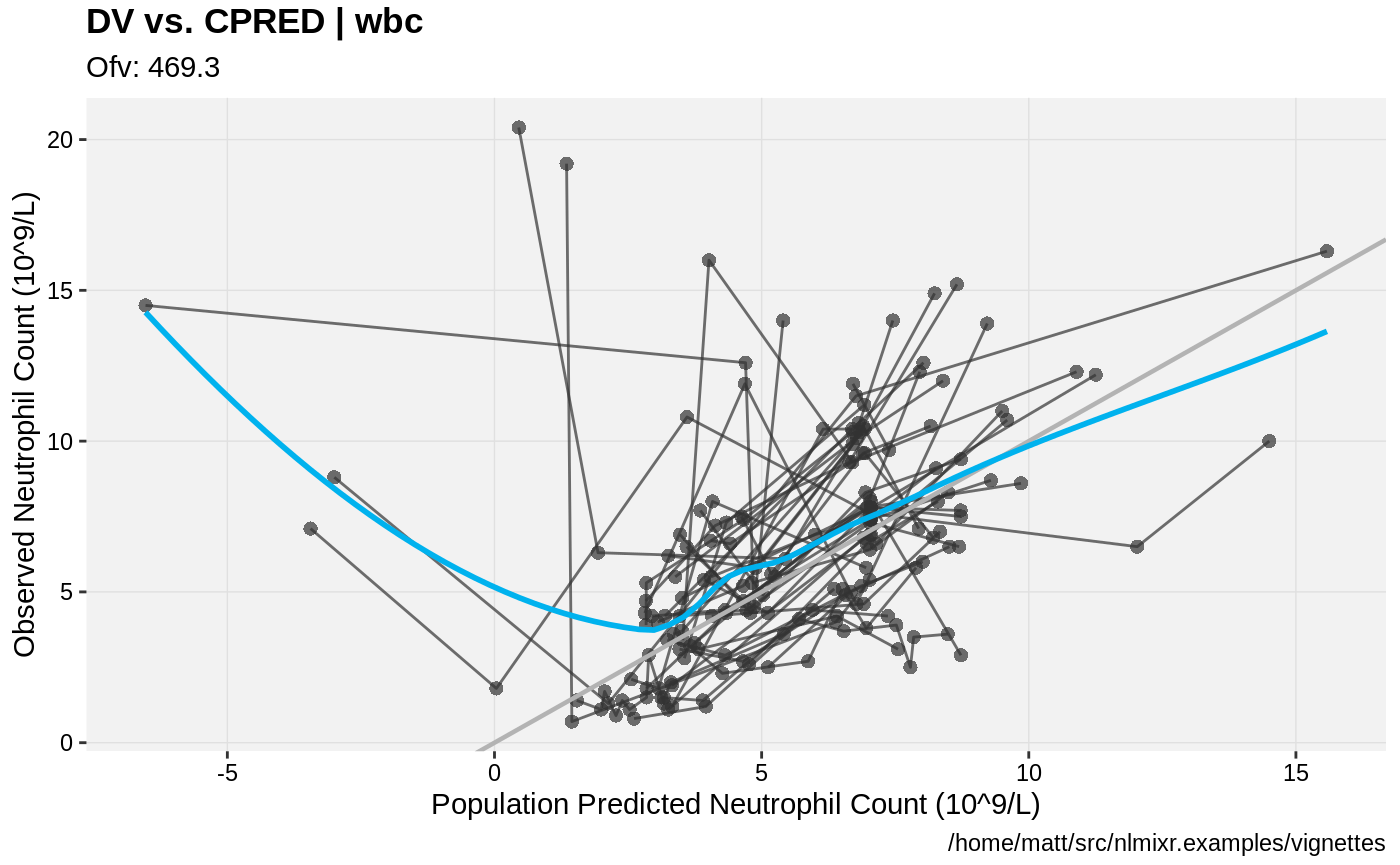
print(dv_vs_pred(xpdb) +
ylab("Observed Neutrophil Count (10^9/L)") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))
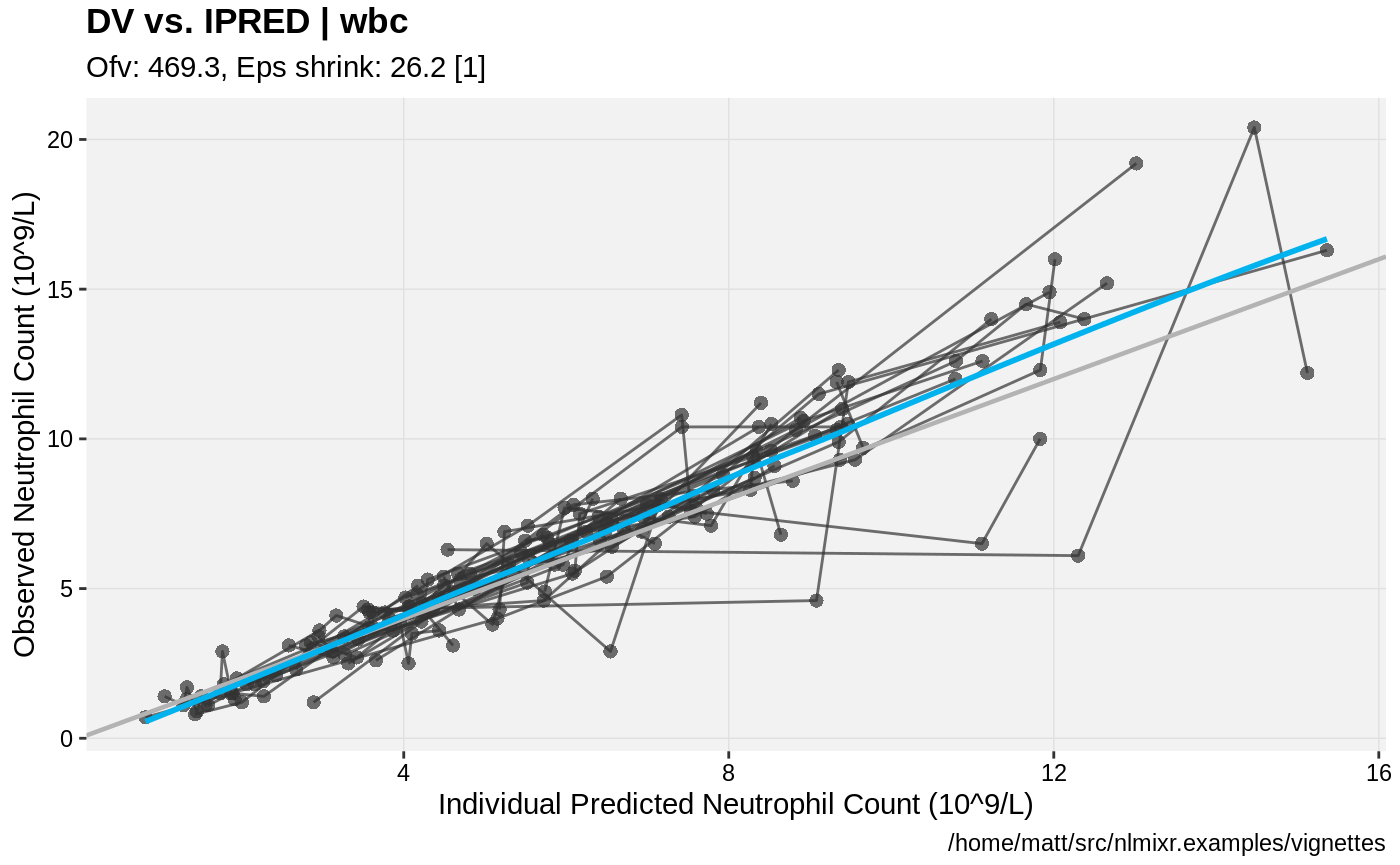
print(dv_vs_ipred(xpdb) +
ylab("Observed Neutrophil Count (10^9/L)") +
xlab("Individual Predicted Neutrophil Count (10^9/L)"))
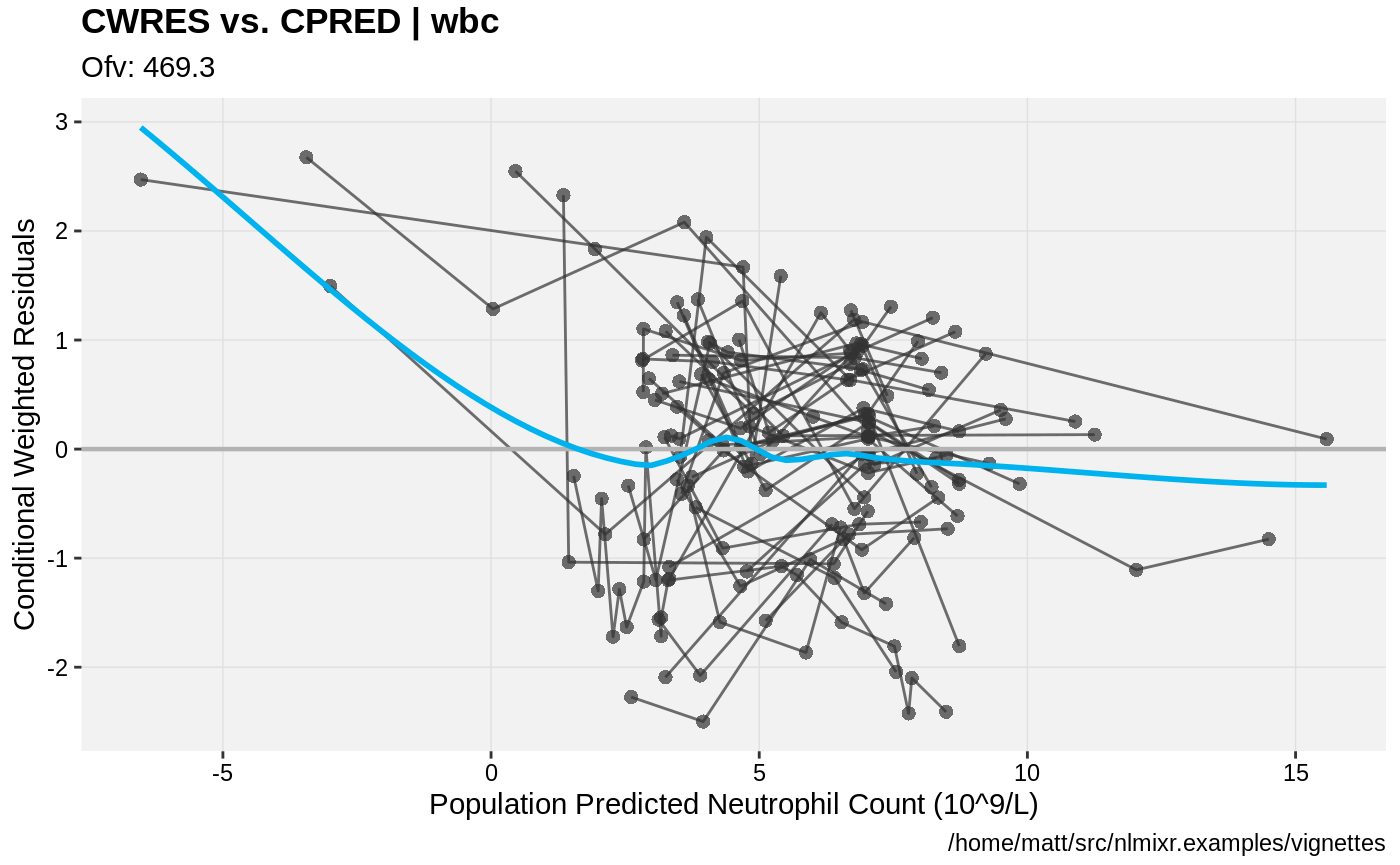
print(res_vs_pred(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))
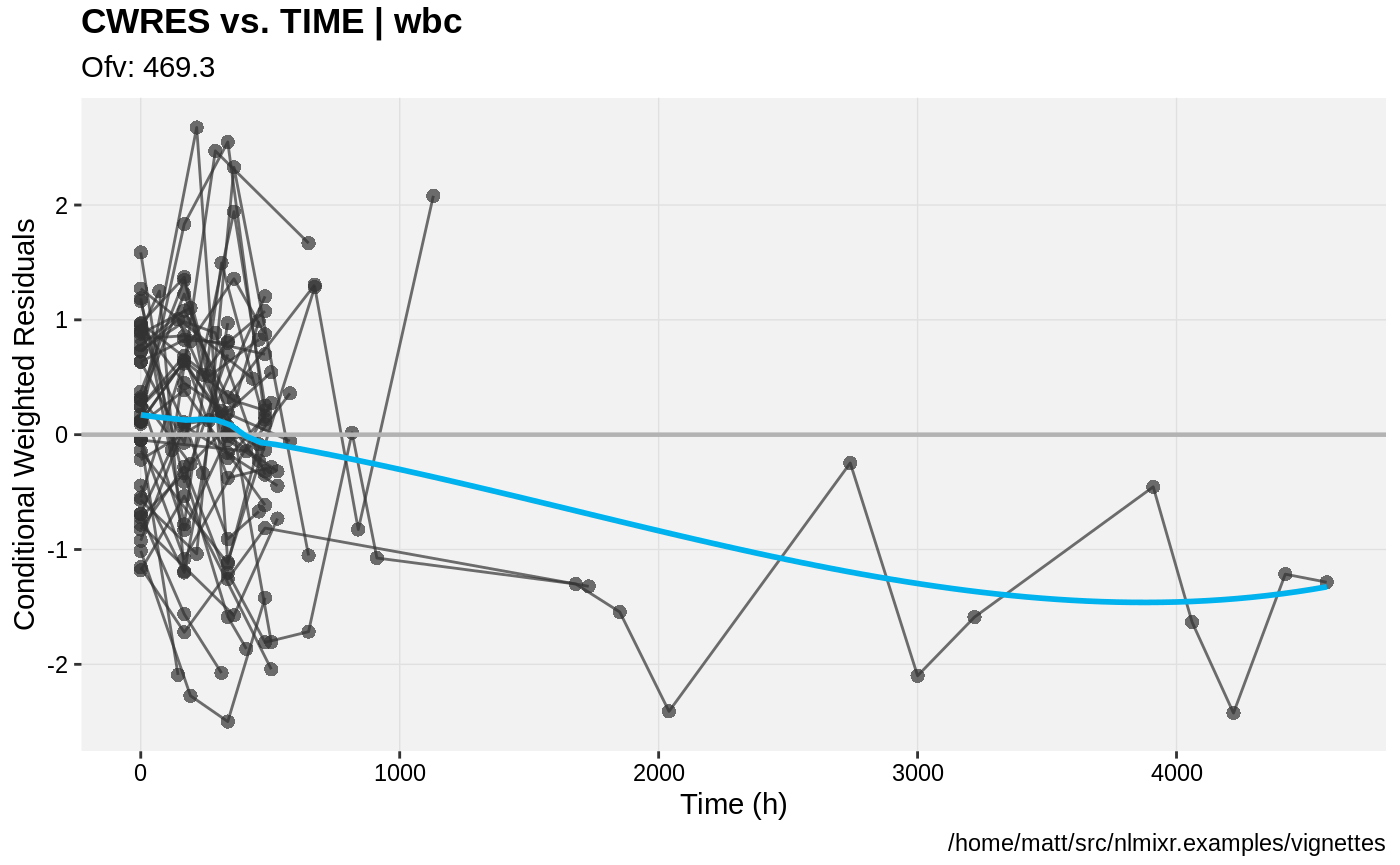
print(res_vs_idv(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Time (h)"))
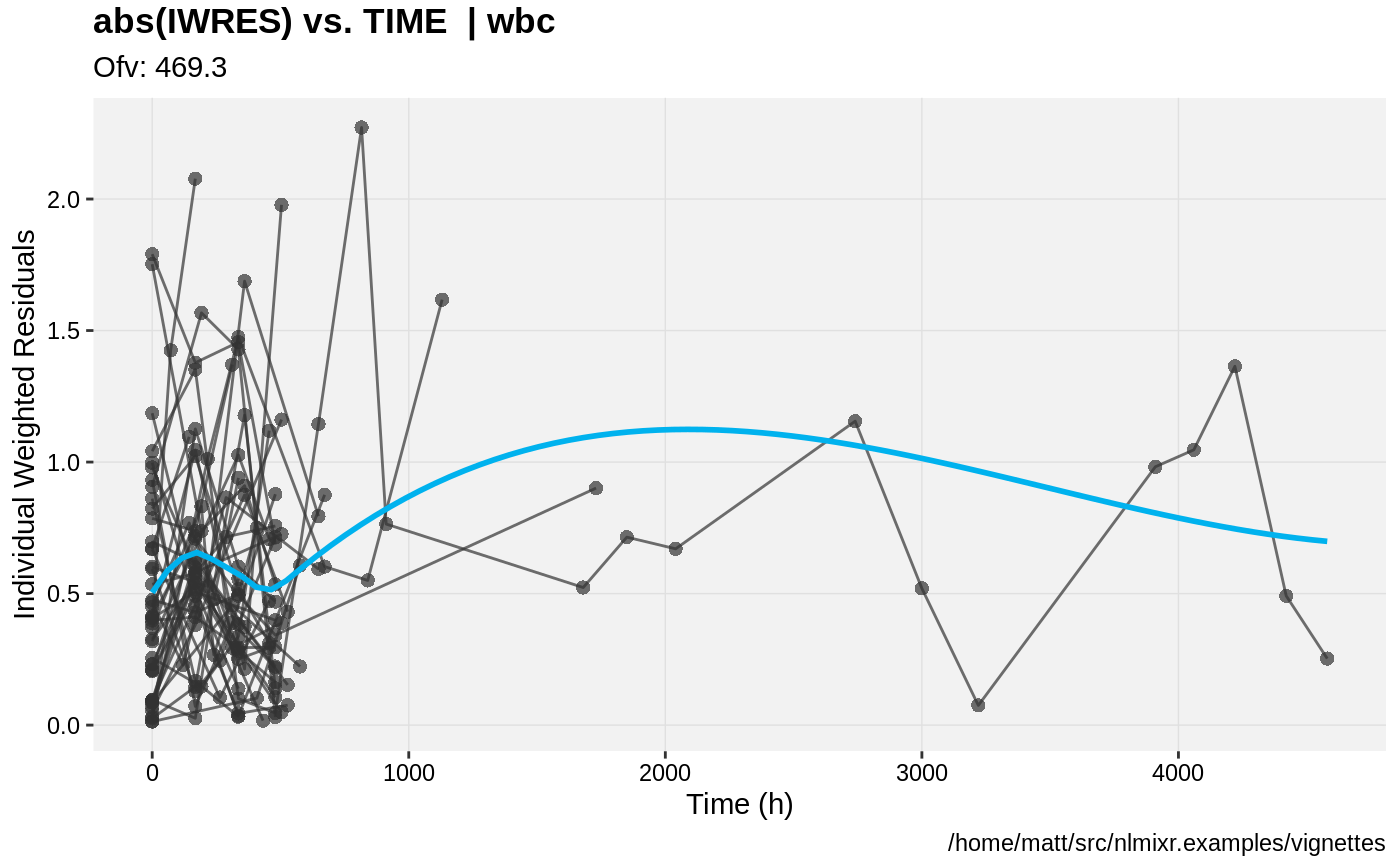
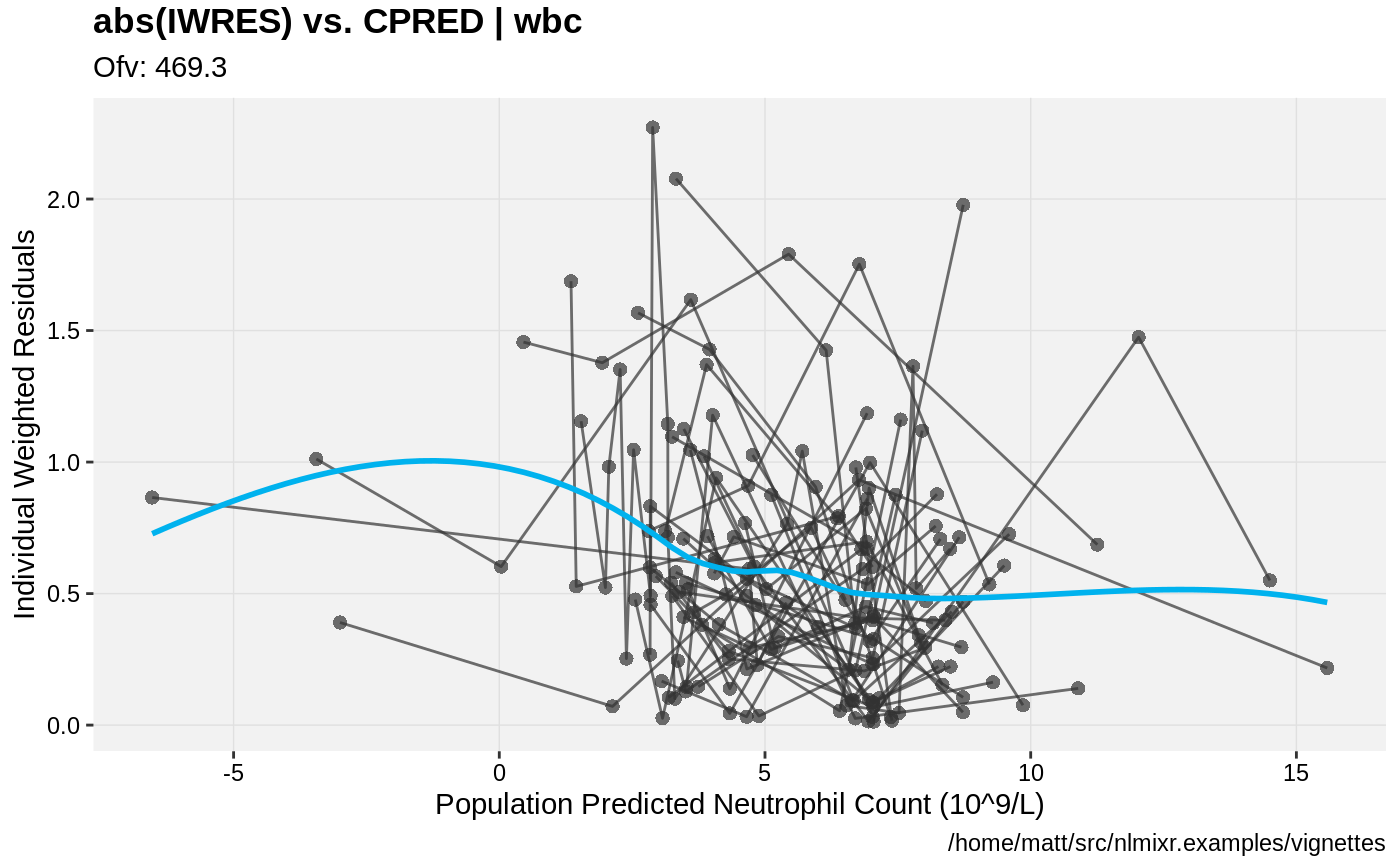
print(absval_res_vs_pred(xpdb, res = 'IWRES') +
ylab("Individual Weighted Residuals") +
xlab("Population Predicted Neutrophil Count (10^9/L)"))
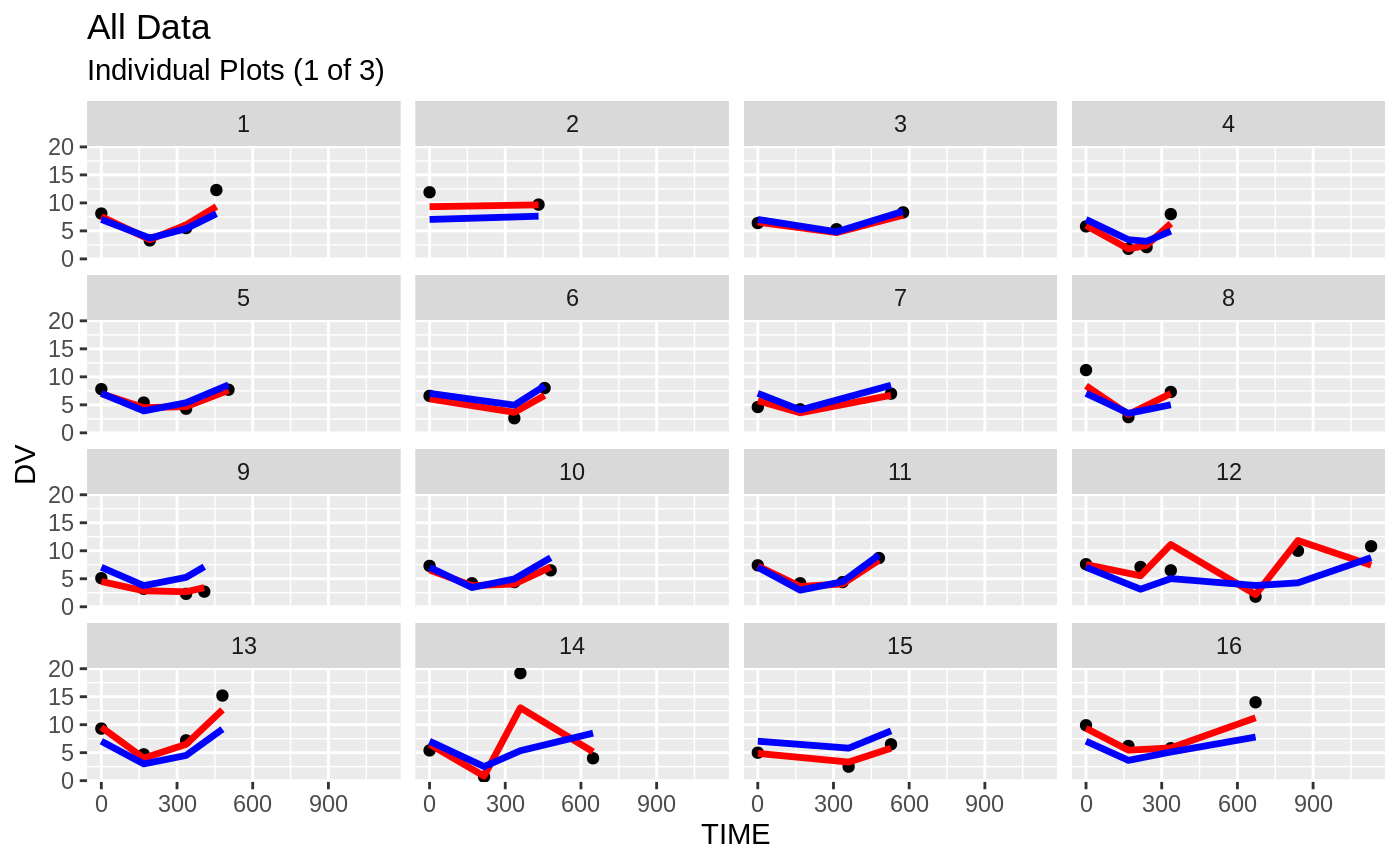
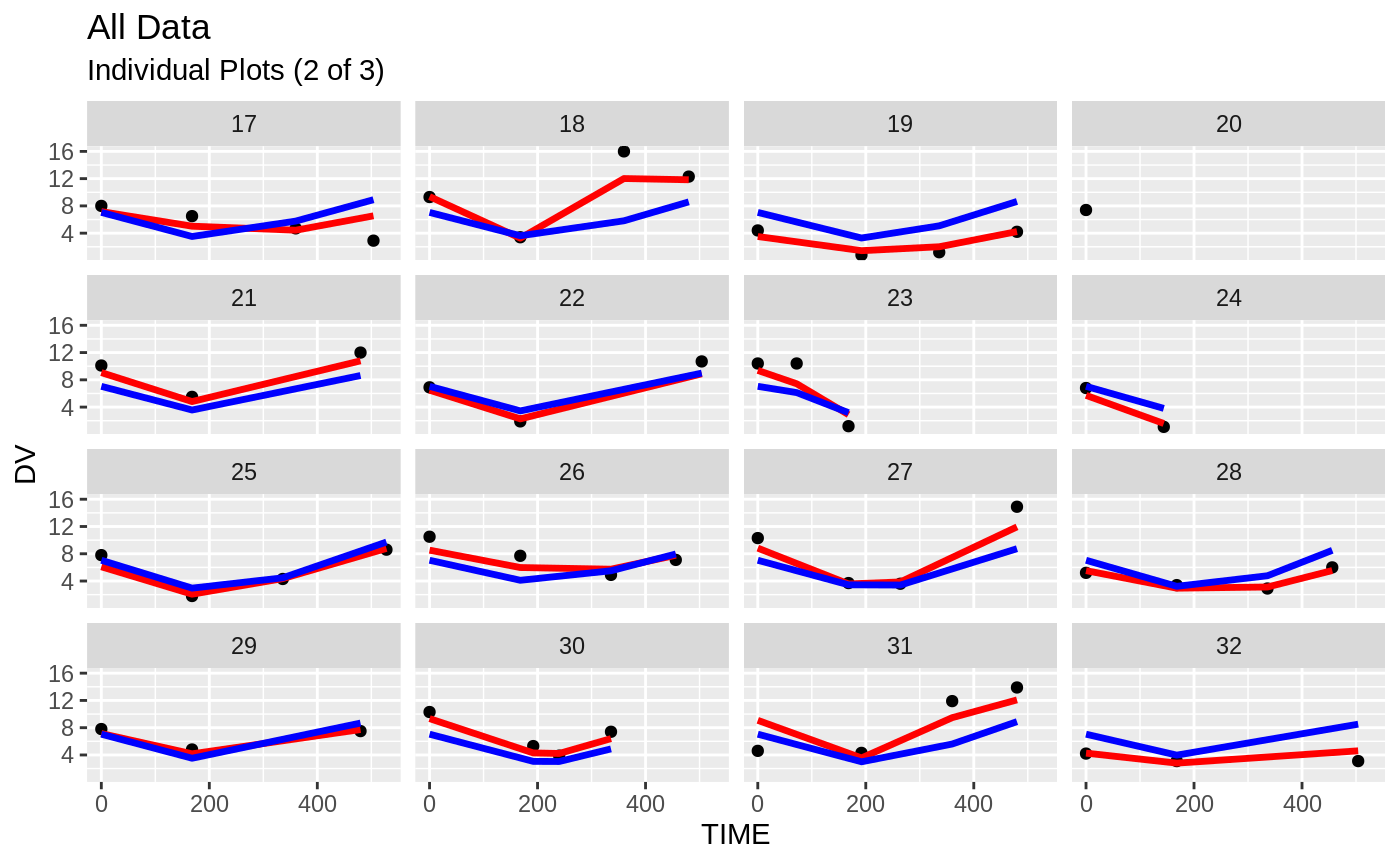
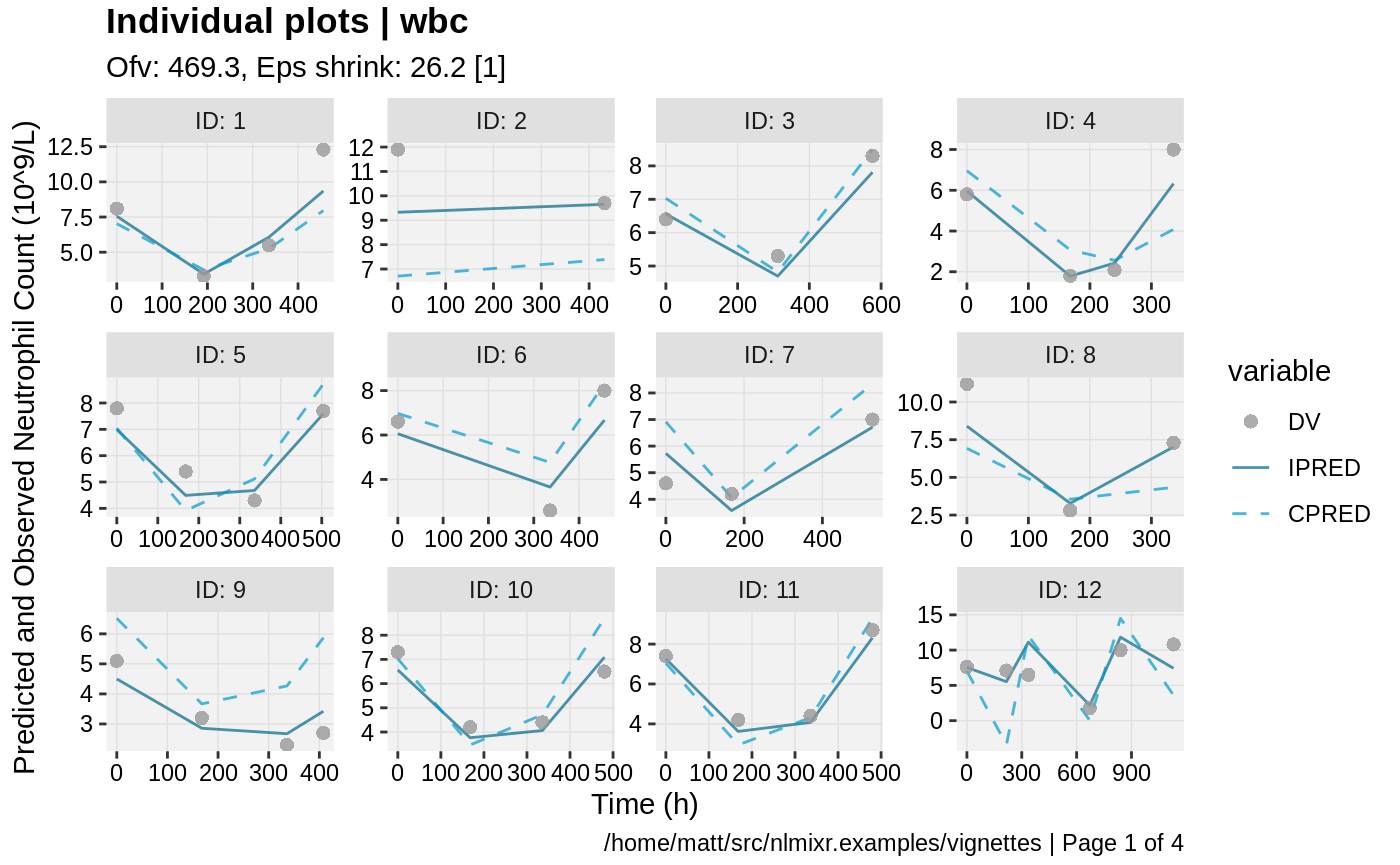
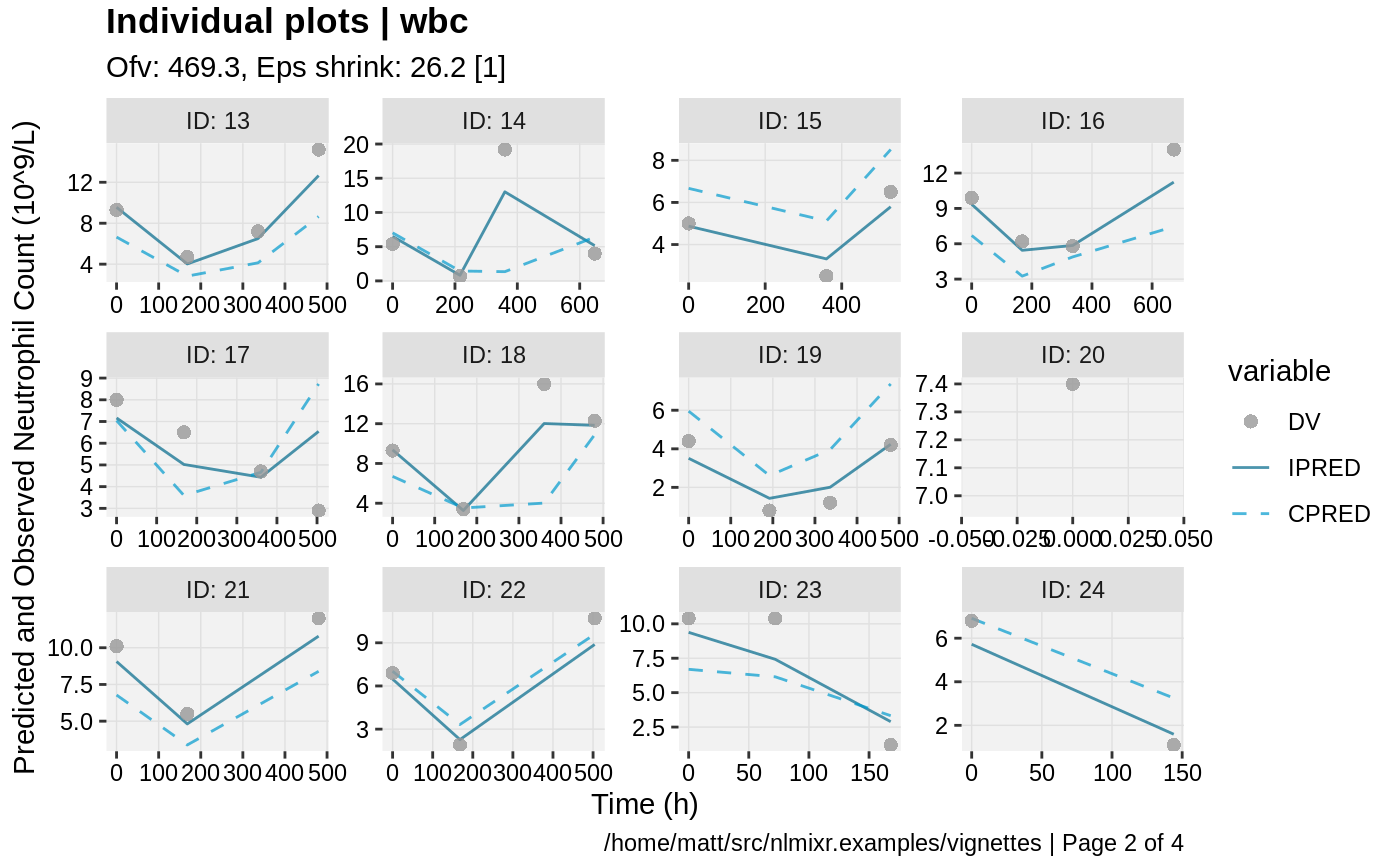
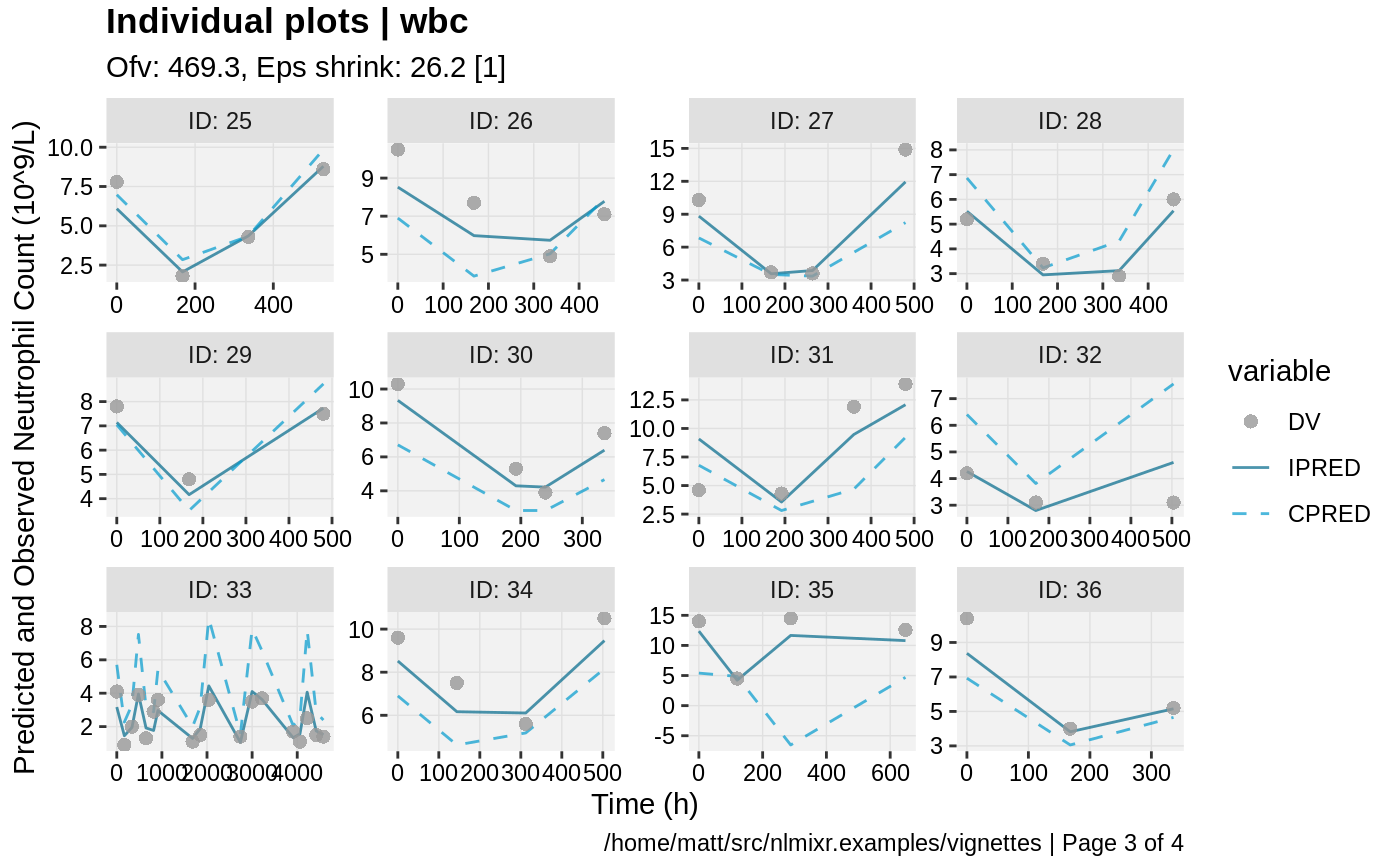
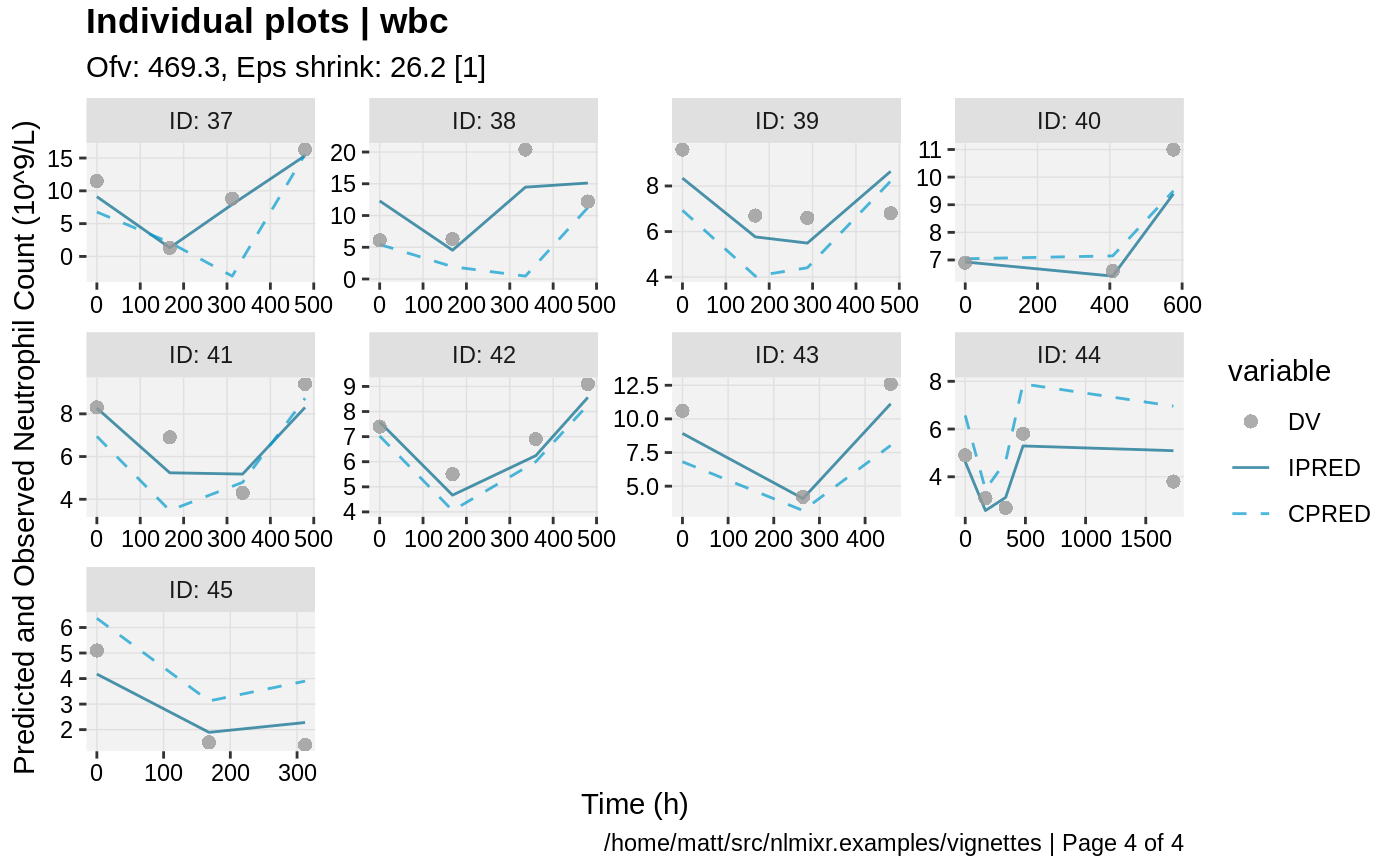
print(ind_plots(xpdb, nrow=3, ncol=4) +
ylab("Predicted and Observed Neutrophil Count (10^9/L)") +
xlab("Time (h)"))
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
#> geom_path: Each group consists of only one observation. Do you need to
#> adjust the group aesthetic?
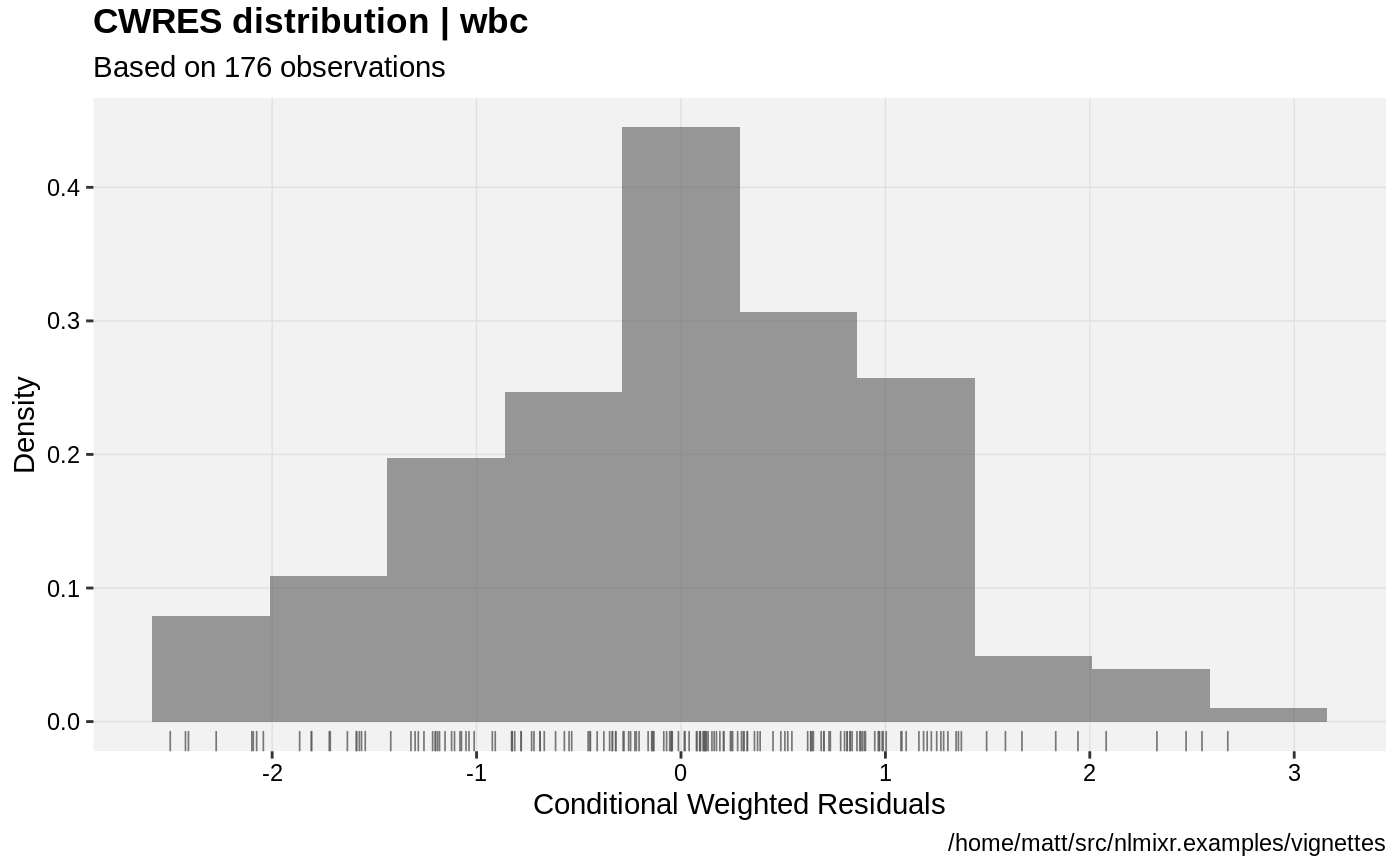
print(res_distrib(xpdb) +
ylab("Density") +
xlab("Conditional Weighted Residuals"))
#vpc.ui(fit.S, n=500, show=list(obs_dv=T), ylab = "Neutrophil count (10^9/L)", xlab = "Time (h)")
# 10 bins is slightly better than auto bin
vpc.ui(fit.F, n=500, n_bins = 10, show=list(obs_dv=T), ylab = "Neutrophil Count (10^9/L)", xlab = "Time (h)")
#> Loading vpc already run (/tmp/RtmpXKdUFj/temp_libpath58be457a881a/nlmixr.examples/nlmixr-vpc-wbc-d3--c2e7b79f91651851066ecd8d902c91ee.rds)
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now
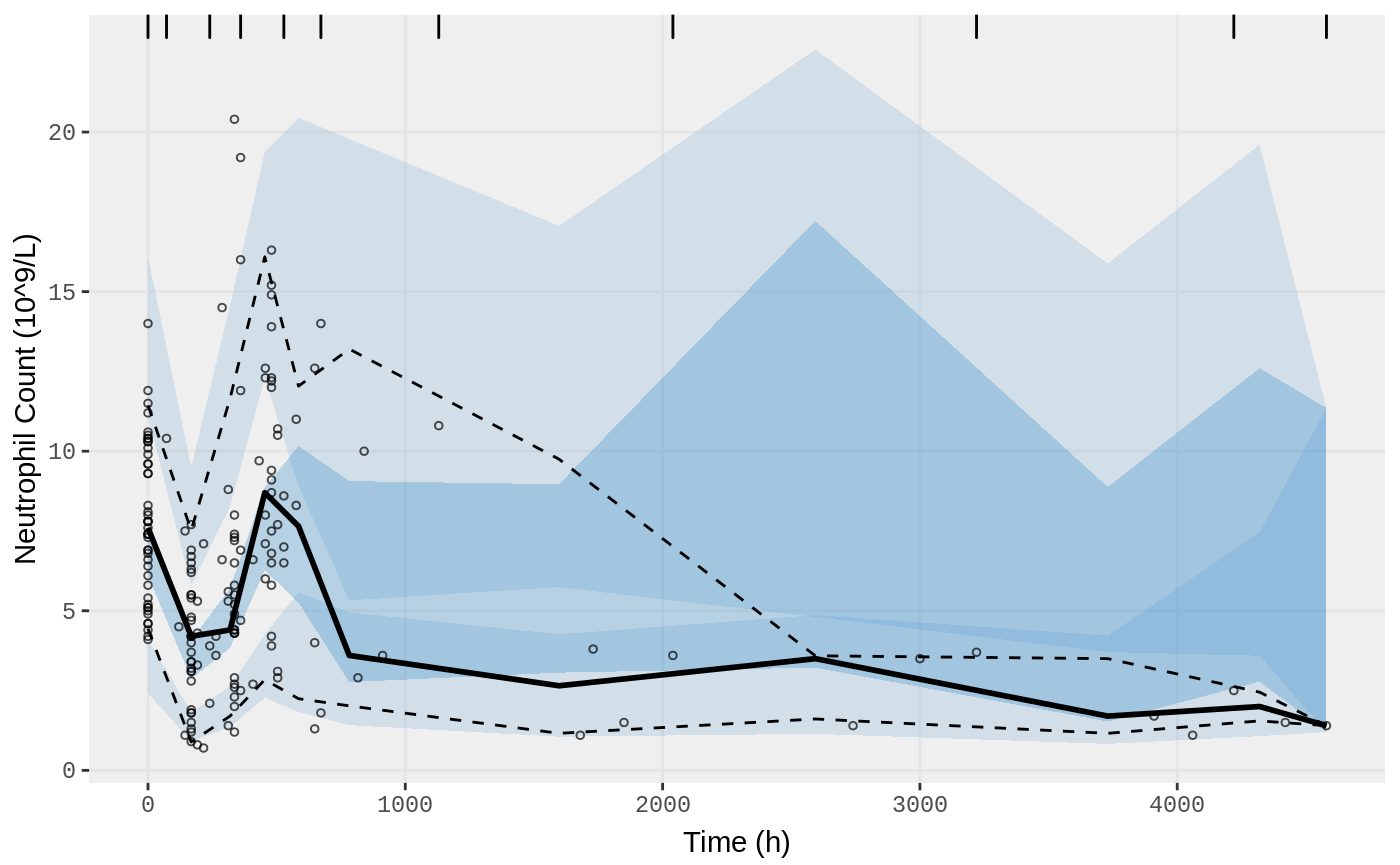
# specify bins
vpc.ui(fit.F, n=500, bins = c(0,170,300,350,500,600,900,3000,4580), show=list(obs_dv=T), ylab = "Neutrophil Count (10^9/L)", xlab = "Time (h)")
#> Loading vpc already run (/tmp/RtmpXKdUFj/temp_libpath58be457a881a/nlmixr.examples/nlmixr-vpc-wbc-d3--bb99abc56c26bc0e6220829df60cd738.rds)
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now