nlmixr
This is an example of a complex model that can be estimated.
In the example below, a target-mediated drug disposition PK model for nimotuzumab is illustrated (Rodríguez-Vera et al. 2015).
Model Schematic
nlmixr model
library(nlmixr)
## Note to get the datasets you can use:
## remotes::install_github("nlmixrdevelopment/nlmixr.examples")
library(nlmixr.examples) ## For nlmixr.examples data
library(xpose)
#> Loading required package: ggplot2
#>
#> Attaching package: 'xpose'
#> The following object is masked from 'package:nlmixr':
#>
#> vpc
#> The following object is masked from 'package:stats':
#>
#> filter
library(xpose.nlmixr)
#>
#> Attaching package: 'xpose.nlmixr'
#> The following object is masked from 'package:nlmixr':
#>
#> vpc
library(ggplot2)
nimo <- function() {
ini({
## Note that the UI can take expressions
## Also note that these initial estimates should be provided on the log-scale
tcl <- log(0.001)
tv1 <- log(1.45)
tQ <- log(0.004)
tv2 <- log(44)
tkss <- log(12)
tkint <- log(0.3)
tksyn <- log(1)
tkdeg <- log(7)
## Initial estimates should be high for SAEM ETAs
eta.cl ~ 2
eta.v1 ~ 2
eta.kss ~ 2
## Also true for additive error (also ignored in SAEM)
add.err <- 10
})
model({
cl <- exp(tcl + eta.cl)
v1 <- exp(tv1 + eta.v1)
Q <- exp(tQ)
v2 <- exp(tv2)
kss <- exp(tkss + eta.kss)
kint <- exp(tkint)
ksyn <- exp(tksyn)
kdeg <- exp(tkdeg)
k <- cl/v1
k12 <- Q/v1
k21 <- Q/v2
eff(0) <- ksyn/kdeg ##initializing compartment
## Concentration is calculated
conc = 0.5*(central/v1-eff-kss)+0.5*sqrt((central/v1-eff-kss)**2+4*kss*central/v1)
d/dt(central) = -(k+k12)*conc*v1+k21*peripheral-kint*eff*conc*v1/(kss+conc)
d/dt(peripheral) = k12*conc*v1-k21*peripheral ##Free Drug second compartment amount
d/dt(eff) = ksyn - kdeg*eff - (kint-kdeg)*conc*eff/(kss+conc)
IPRED=log(conc)
IPRED ~ add(add.err)
})
}Fit
load("nimoData.rda")
fit <- nlmixr(nimo, nimoData, est="saem", control=list(print=0),
table=list(cwres=TRUE, npde=TRUE))
#> Loading model already run (/tmp/RtmpXKdUFj/temp_libpath58be457a881a/nlmixr.examples/nlmixr-nimo-nimoData-saem-1c85d7f280783584155ad6862394fe4b.rds)
#> Warning in lapply(X = X, FUN = FUN, ...): ID missing in parameters dataset;
#> Parameters are assumed to have the same order as the IDs in the event dataset
#> Warning in lapply(X = X, FUN = FUN, ...): Calculated residuals like IPRED
#> are masked by nlmixr calculated valuesGoodness of fit Plots
## Add cwres/npde after fit
fit <- fit %>% addCwres() %>% addNpde();
#> Warning in addNpde(.): Already contains NPDE
## Since it is already there it doesn't actually change anything.
##Goodness-of-fit plots
plot(fit); ## Standard nlmixr plots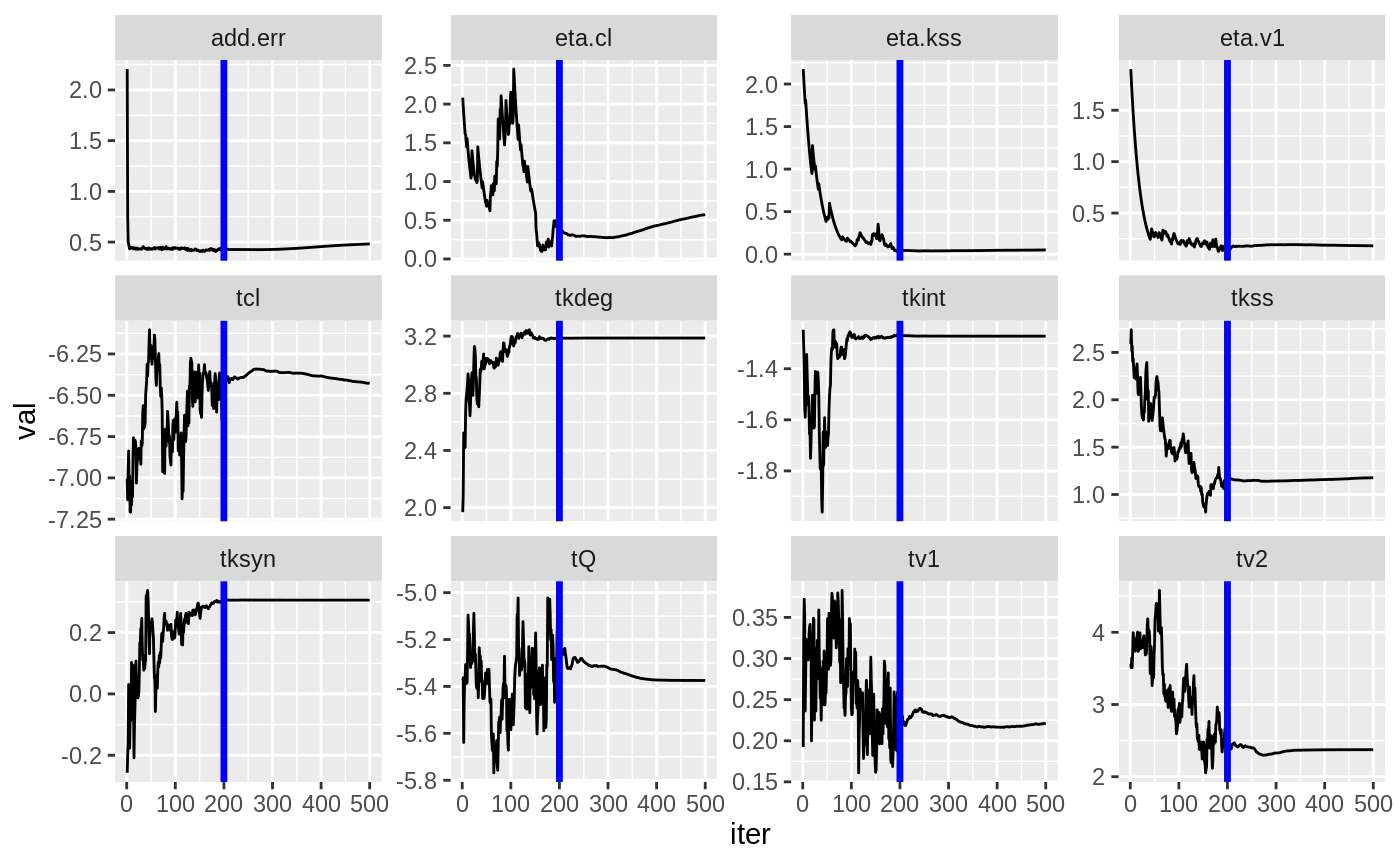
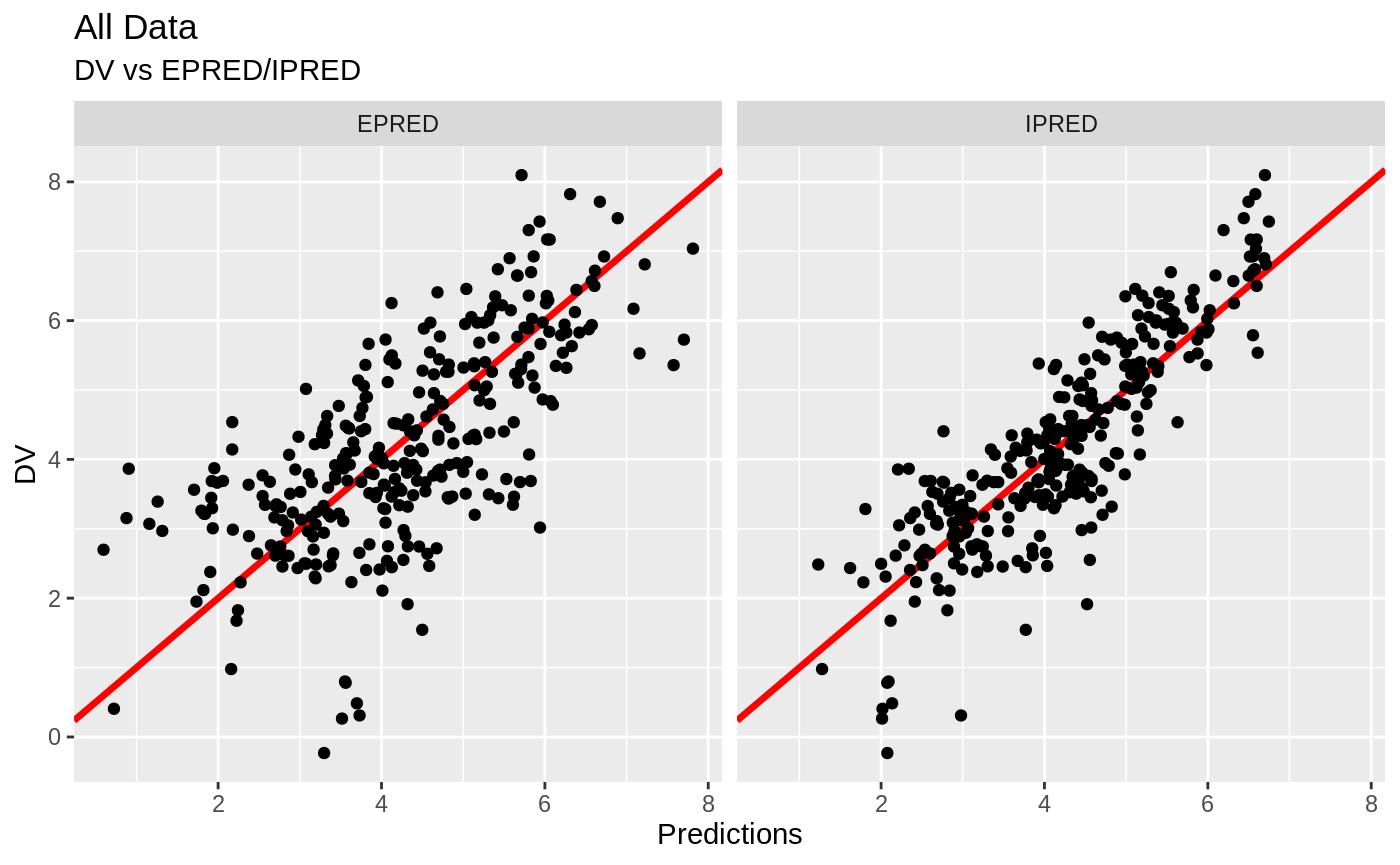
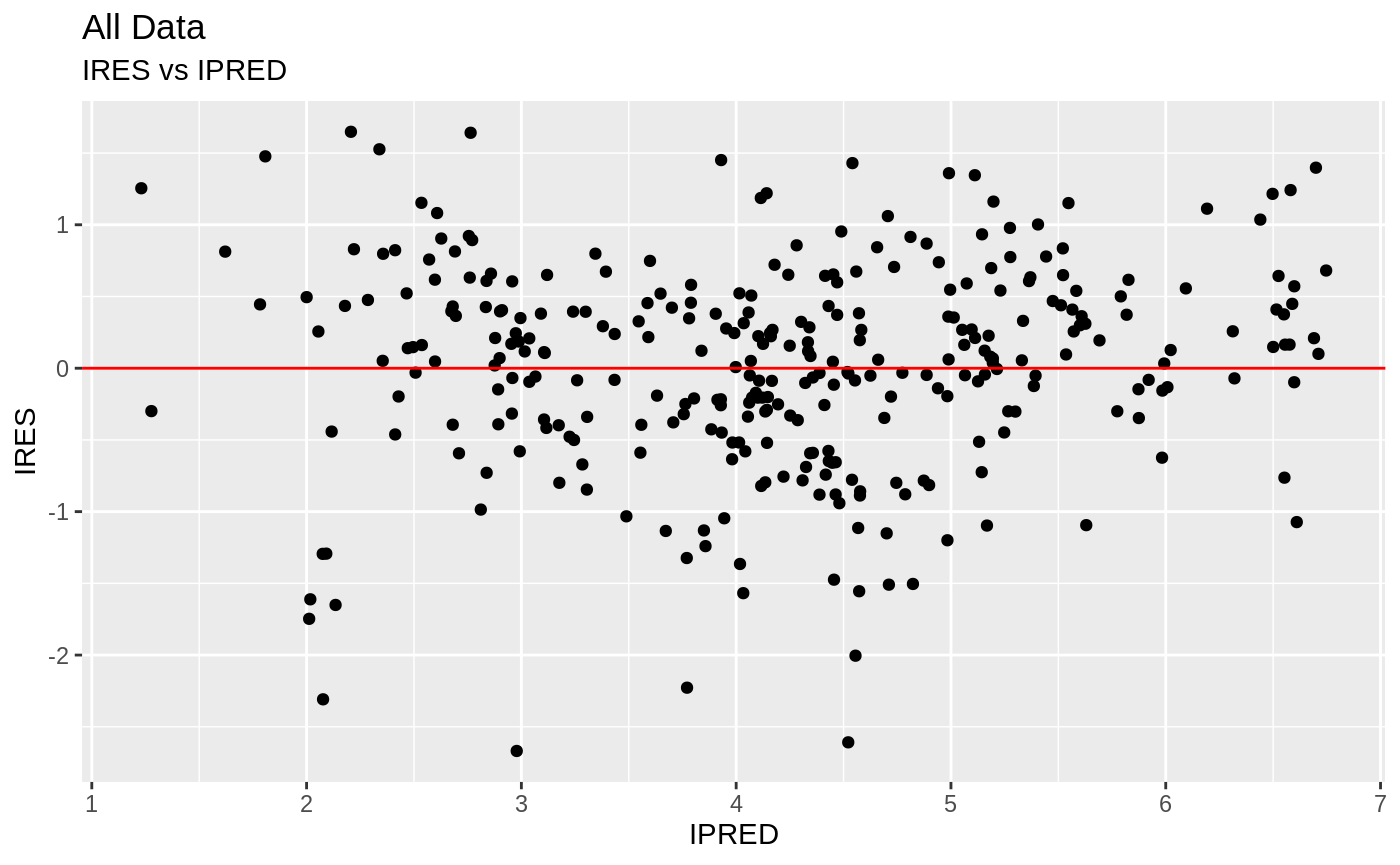
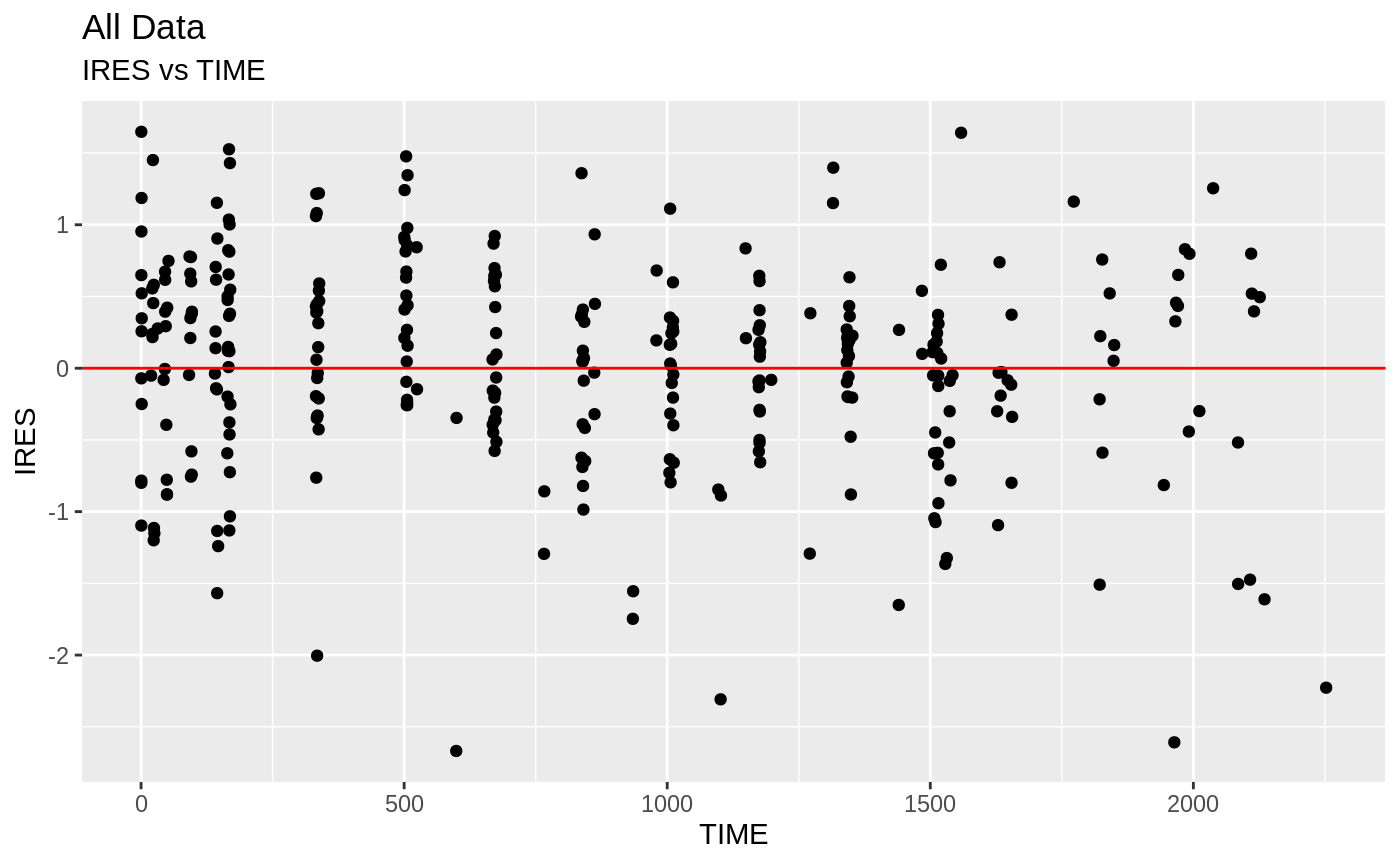
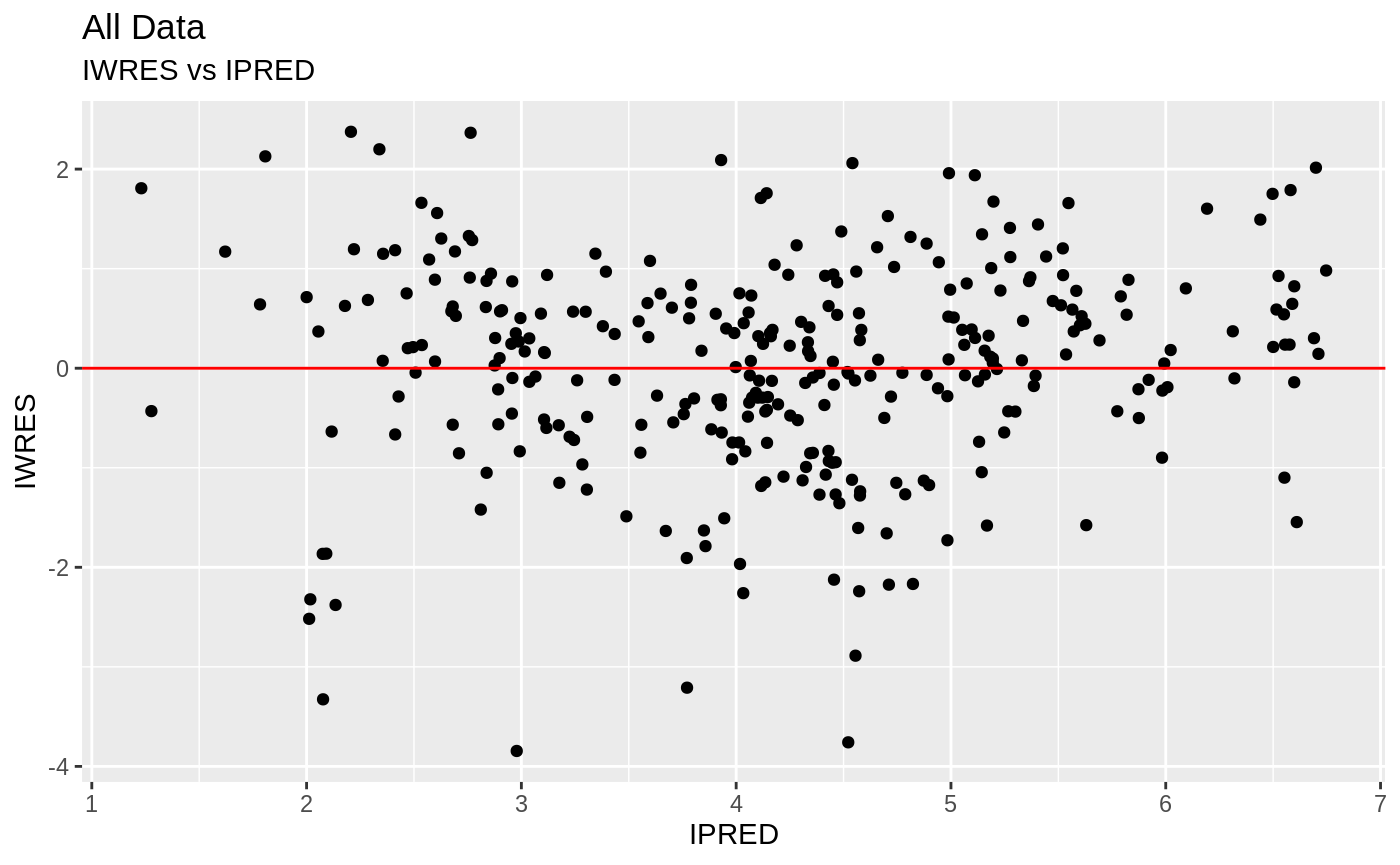
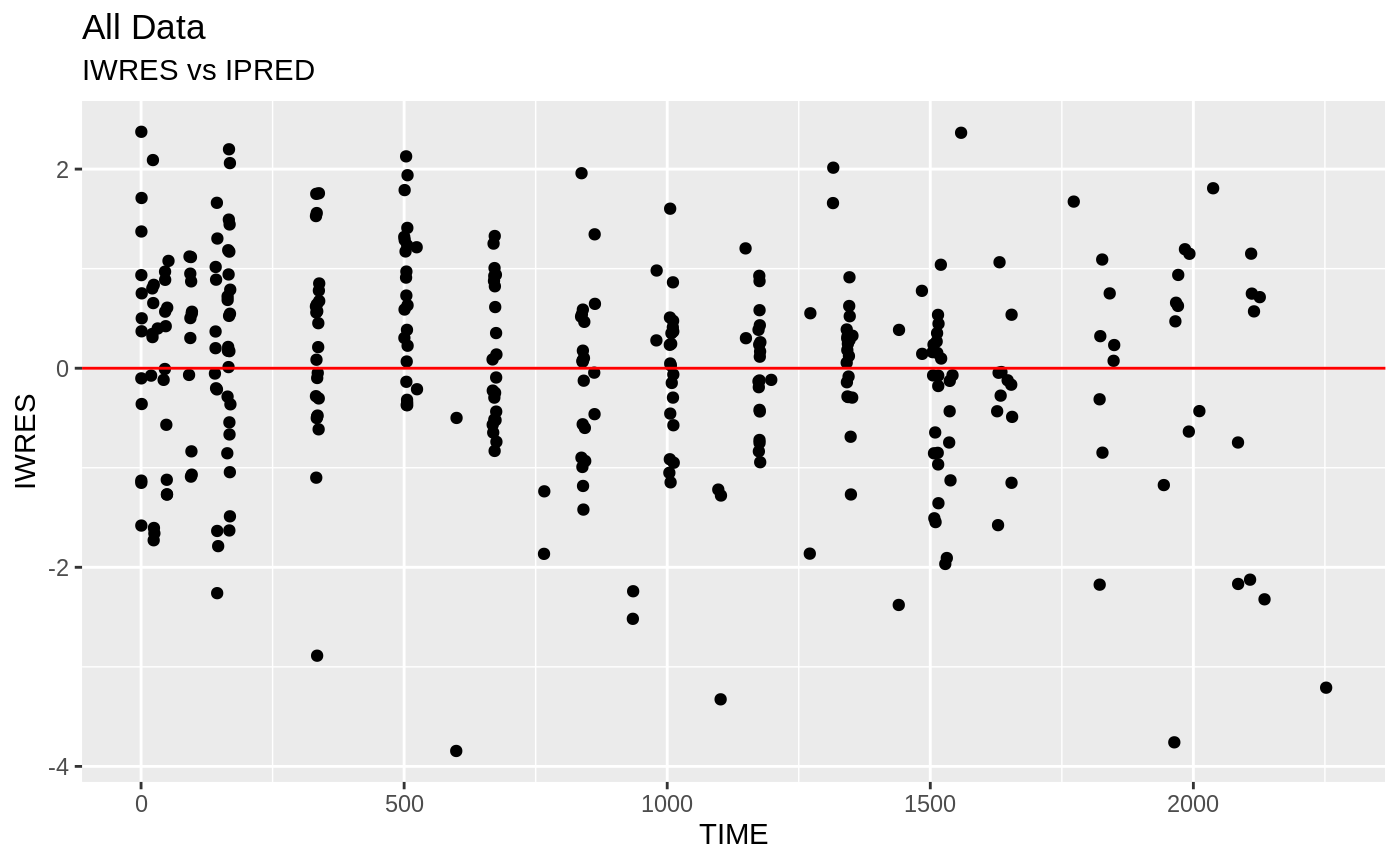
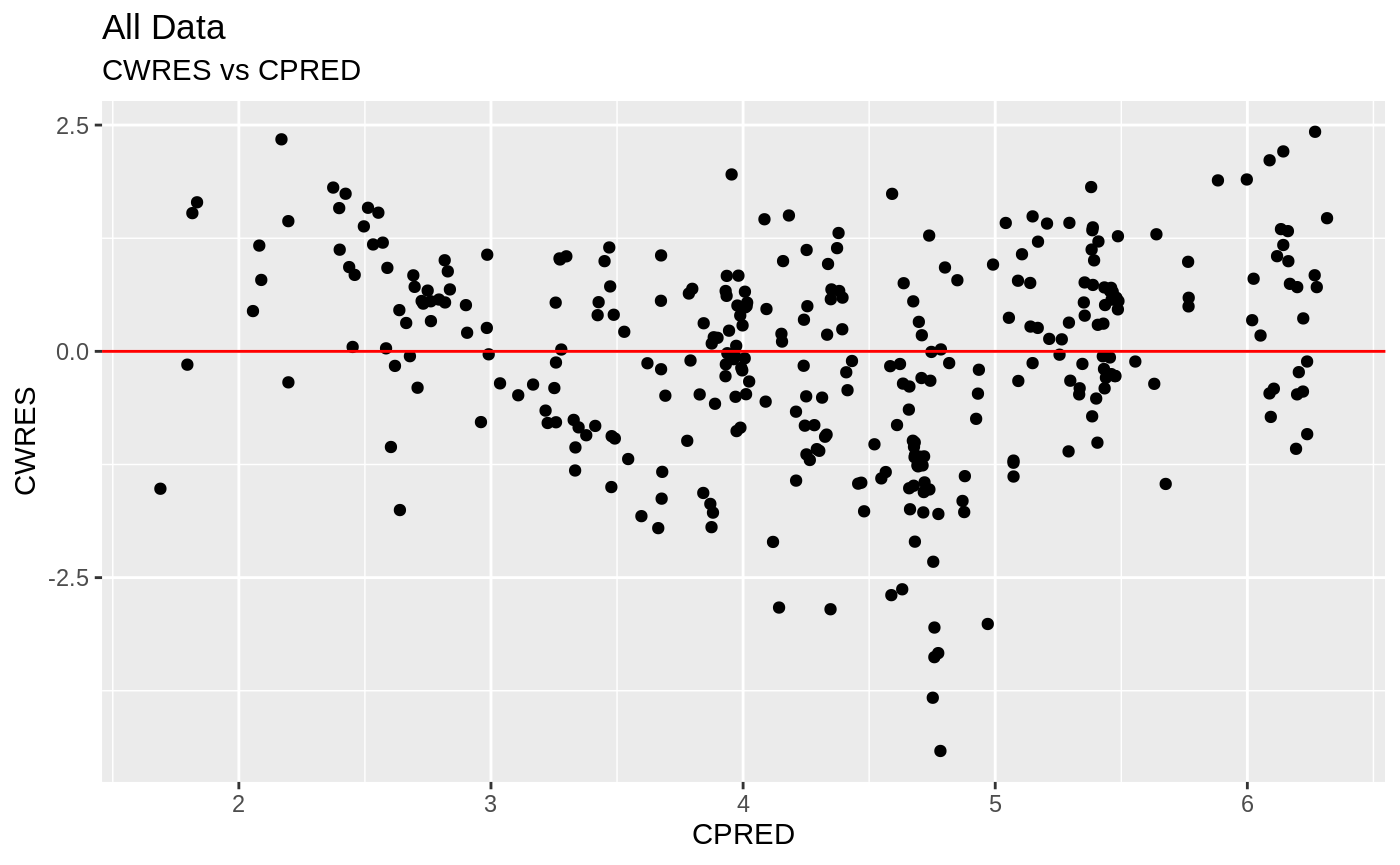
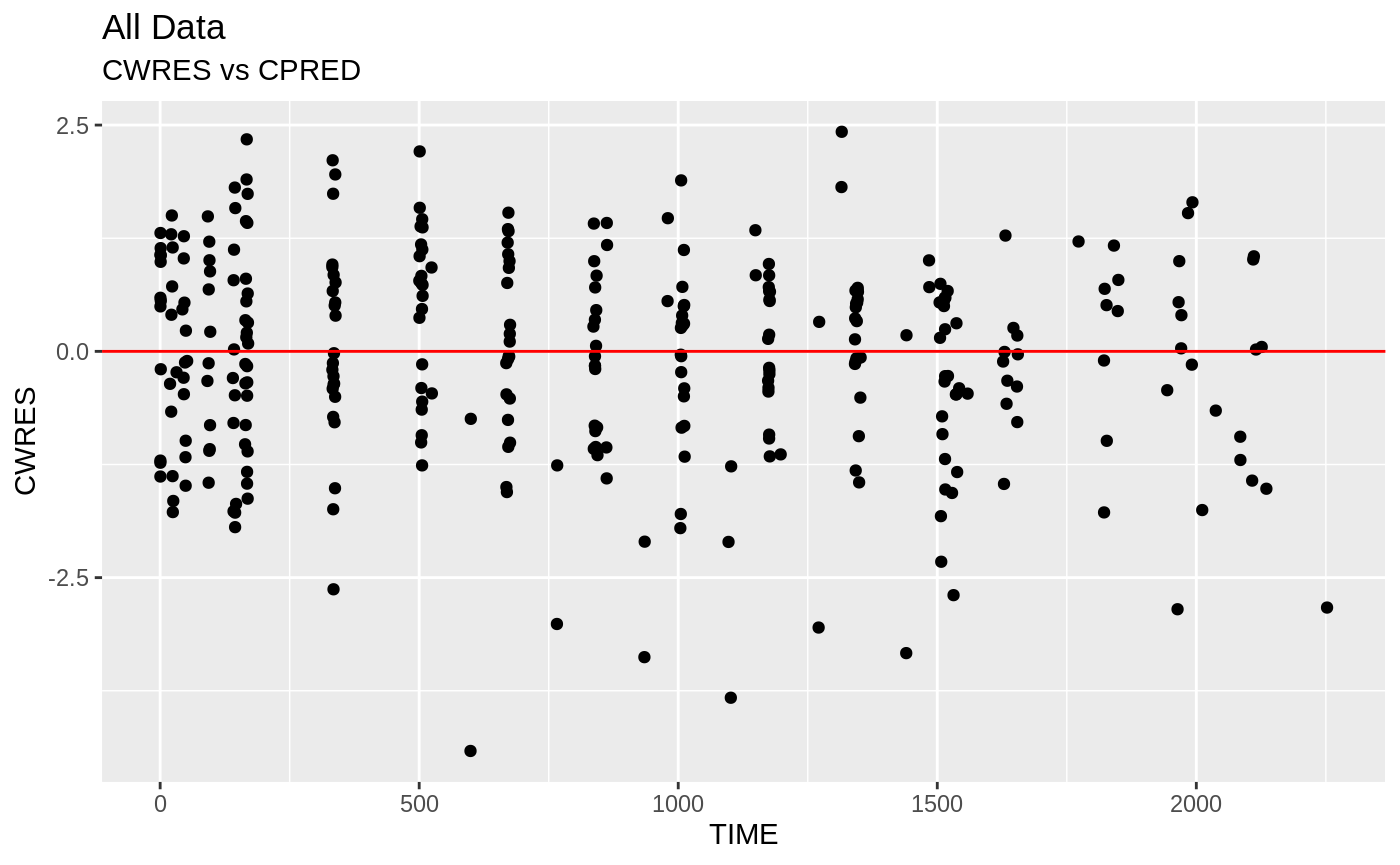
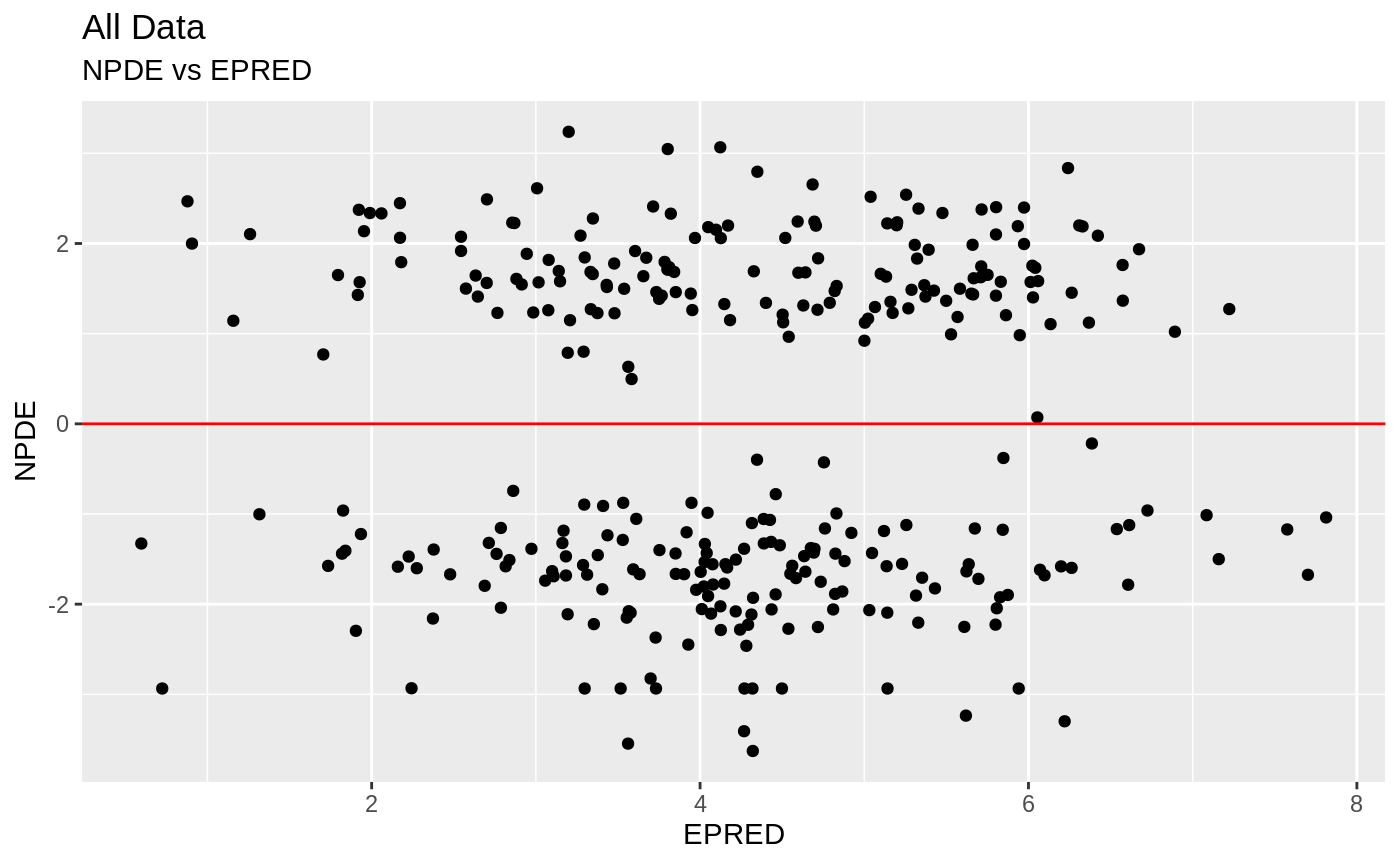
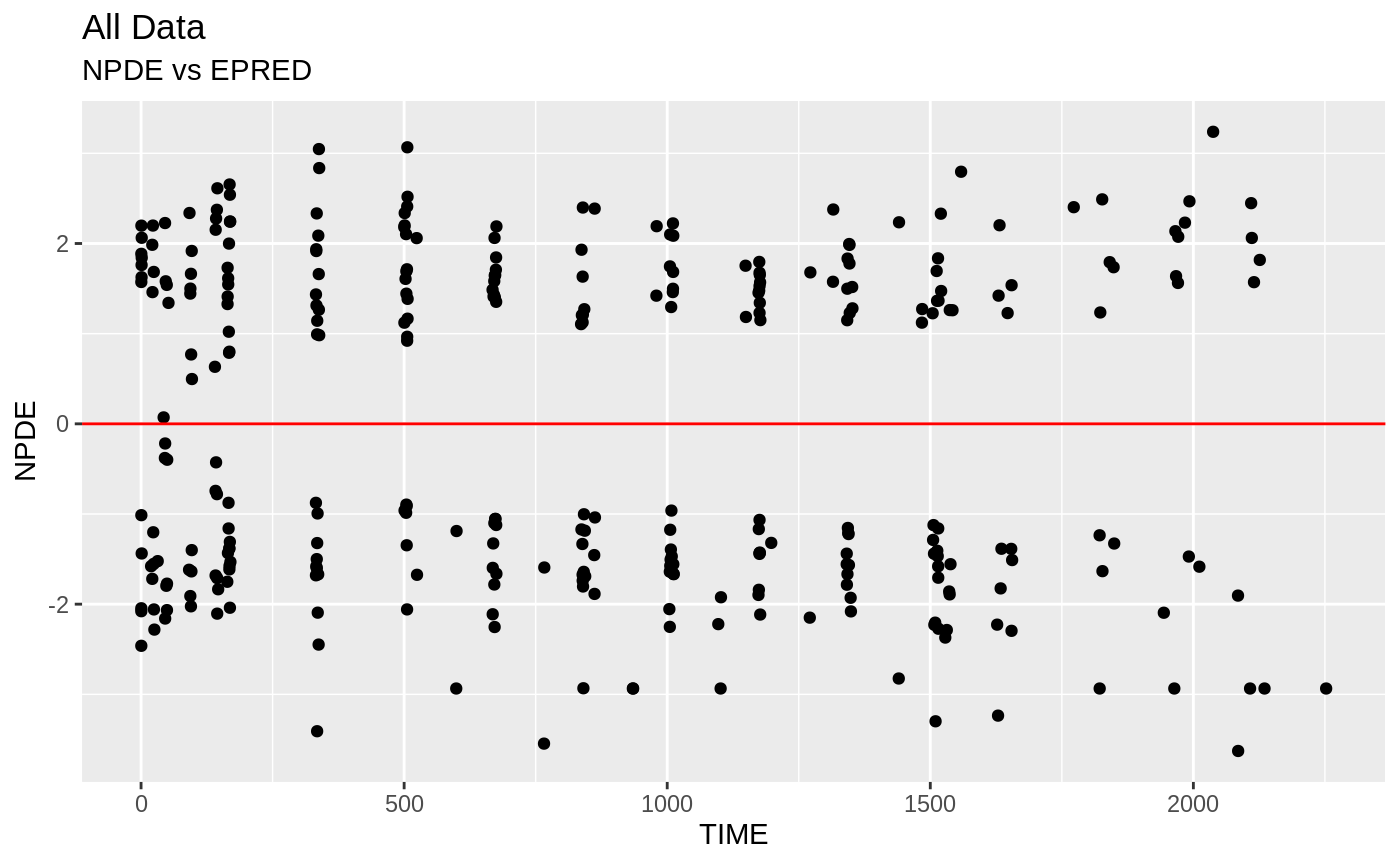
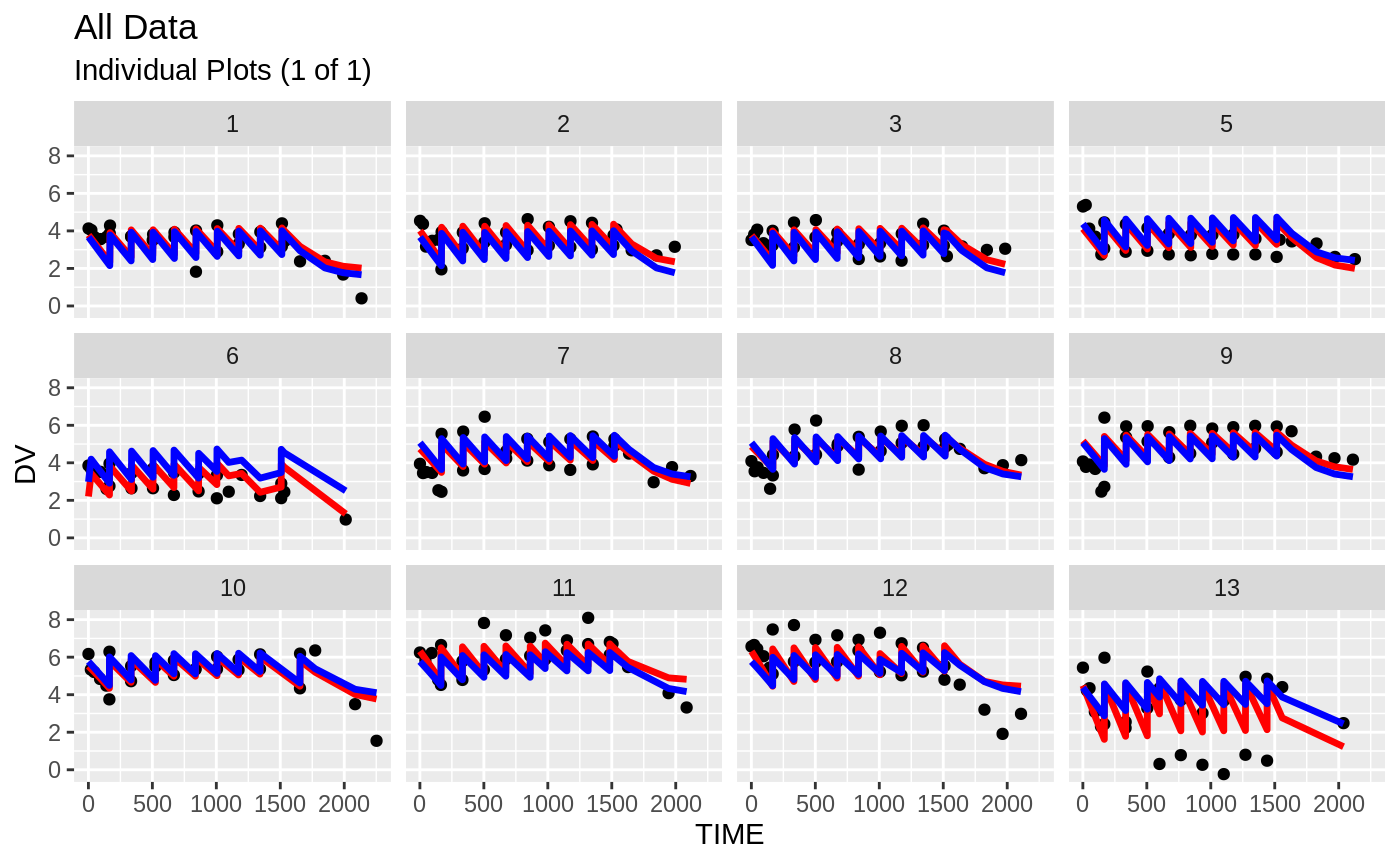
################################################################################
## Xpose plots; Need to print otherwise running a script won't
## show xpose plots
################################################################################
xpdb <- xpose_data_nlmixr(fit) ## first convert to nlmixr object
#> Warning: `complete_()` is deprecated as of tidyr 1.0.0.
#> Please use `complete()` instead.
#> This warning is displayed once per session.
#> Call `lifecycle::last_warnings()` to see where this warning was generated.
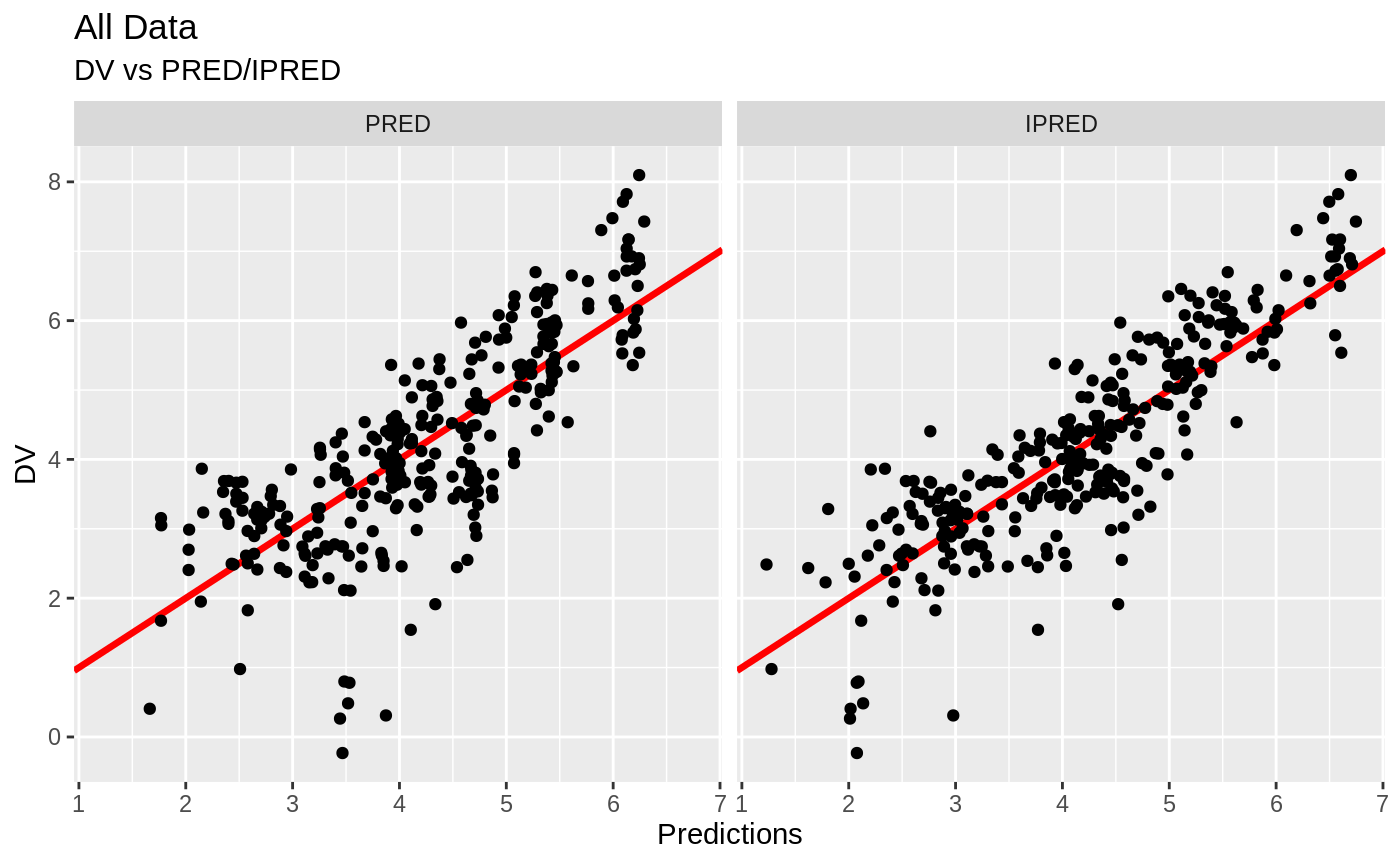
print(dv_vs_pred(xpdb) +
ylab("Observed Nimotuzumab Concentrations (ug/mL)") +
xlab("Population Predicted Nimotuzumab Concentrations (ug/mL)"))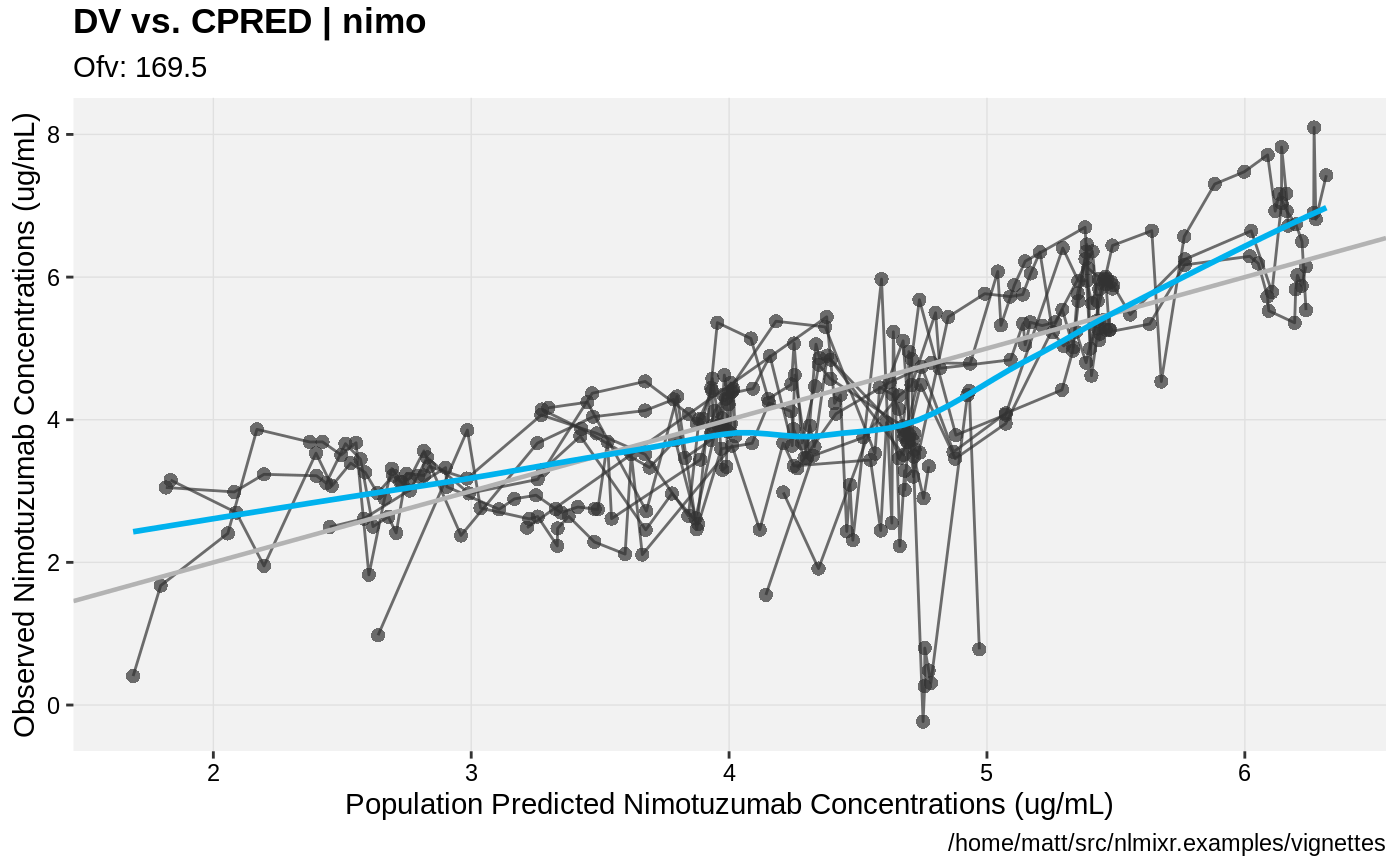
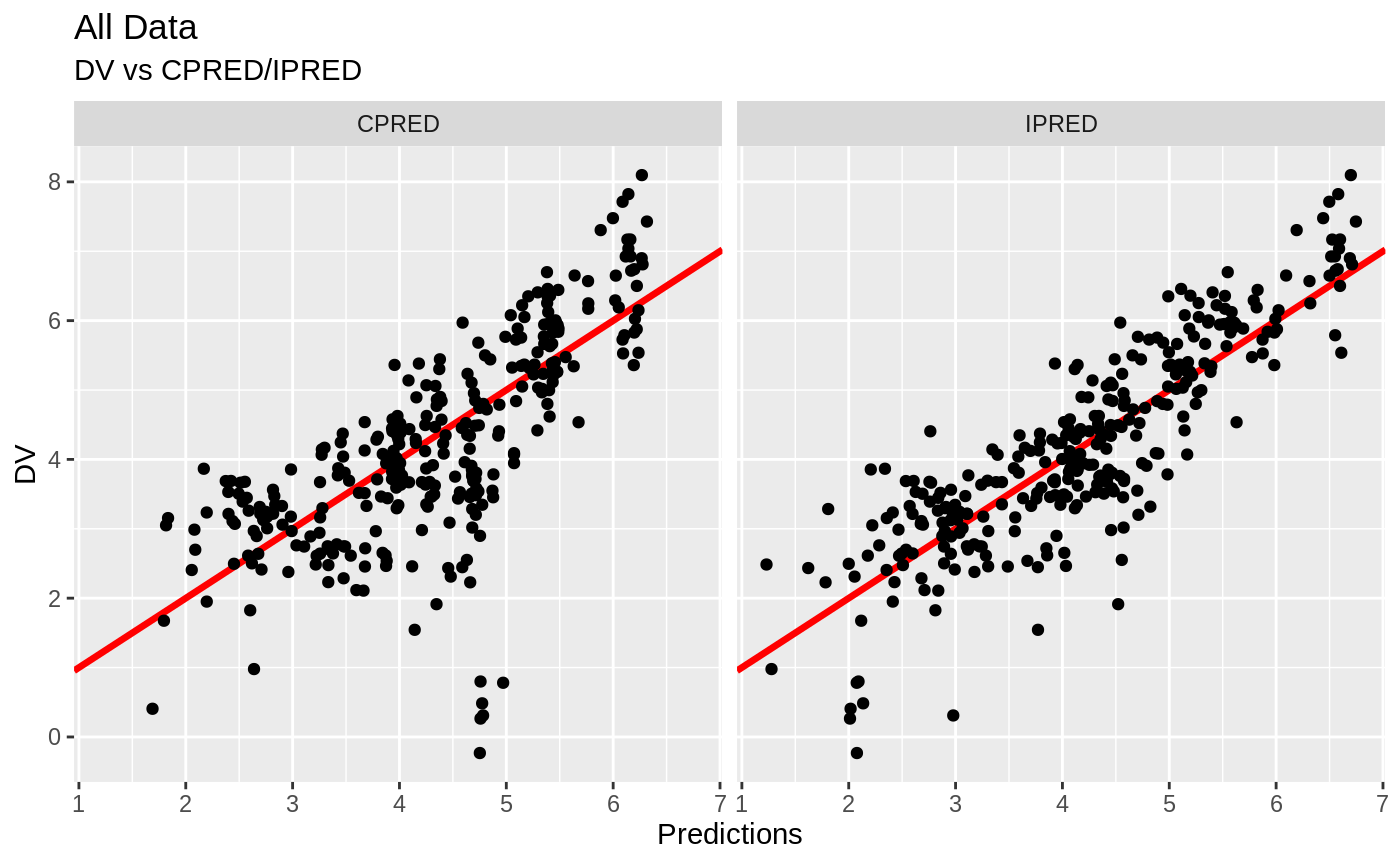
print(dv_vs_ipred(xpdb) +
ylab("Observed Nimotuzumab Concentrations (ug/mL)") +
xlab("Individual Predicted Nimotuzumab Concentrations (ug/mL)"))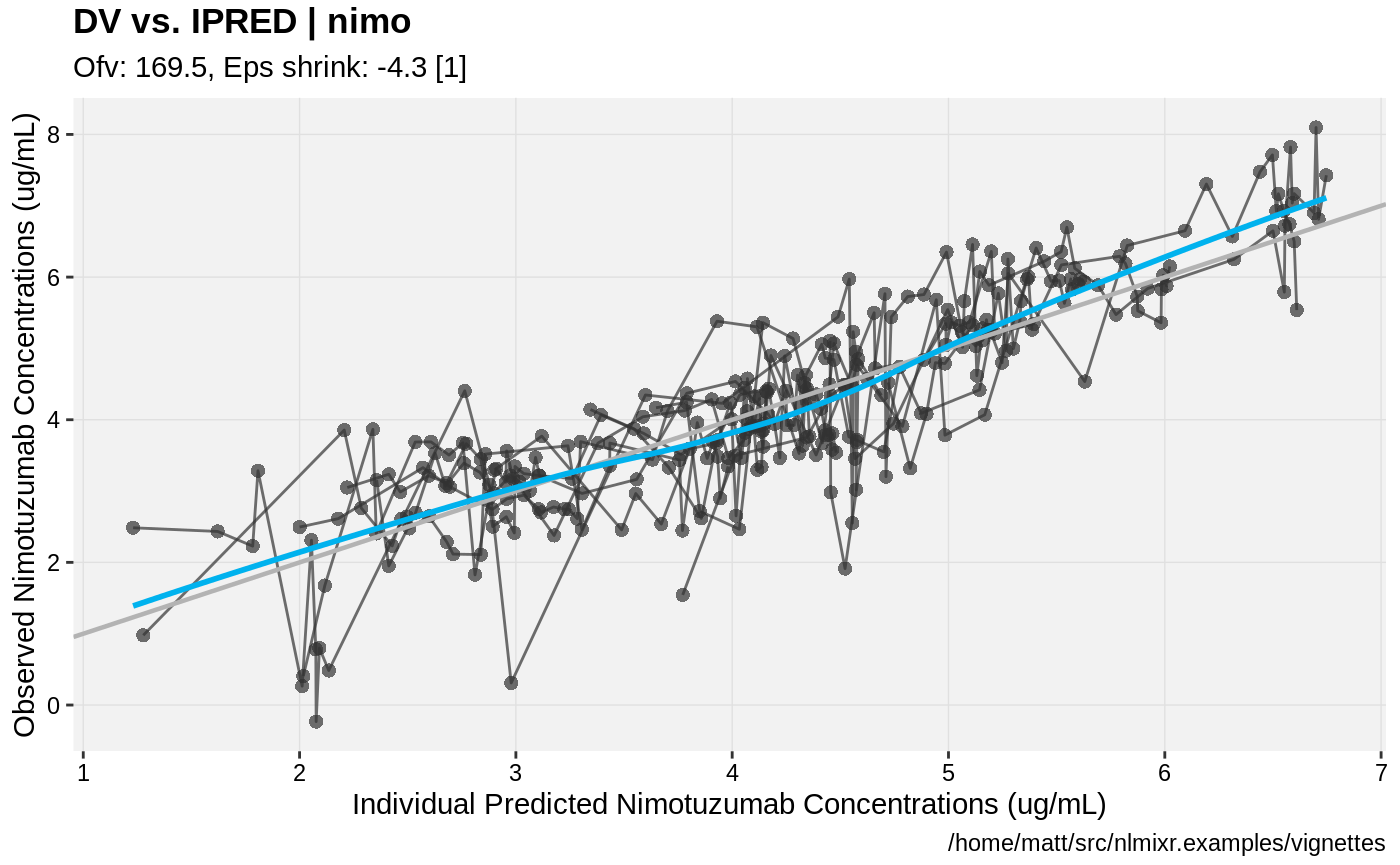
print(res_vs_pred(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Population Predicted Nimotuzumab Concentrations (ug/mL)"))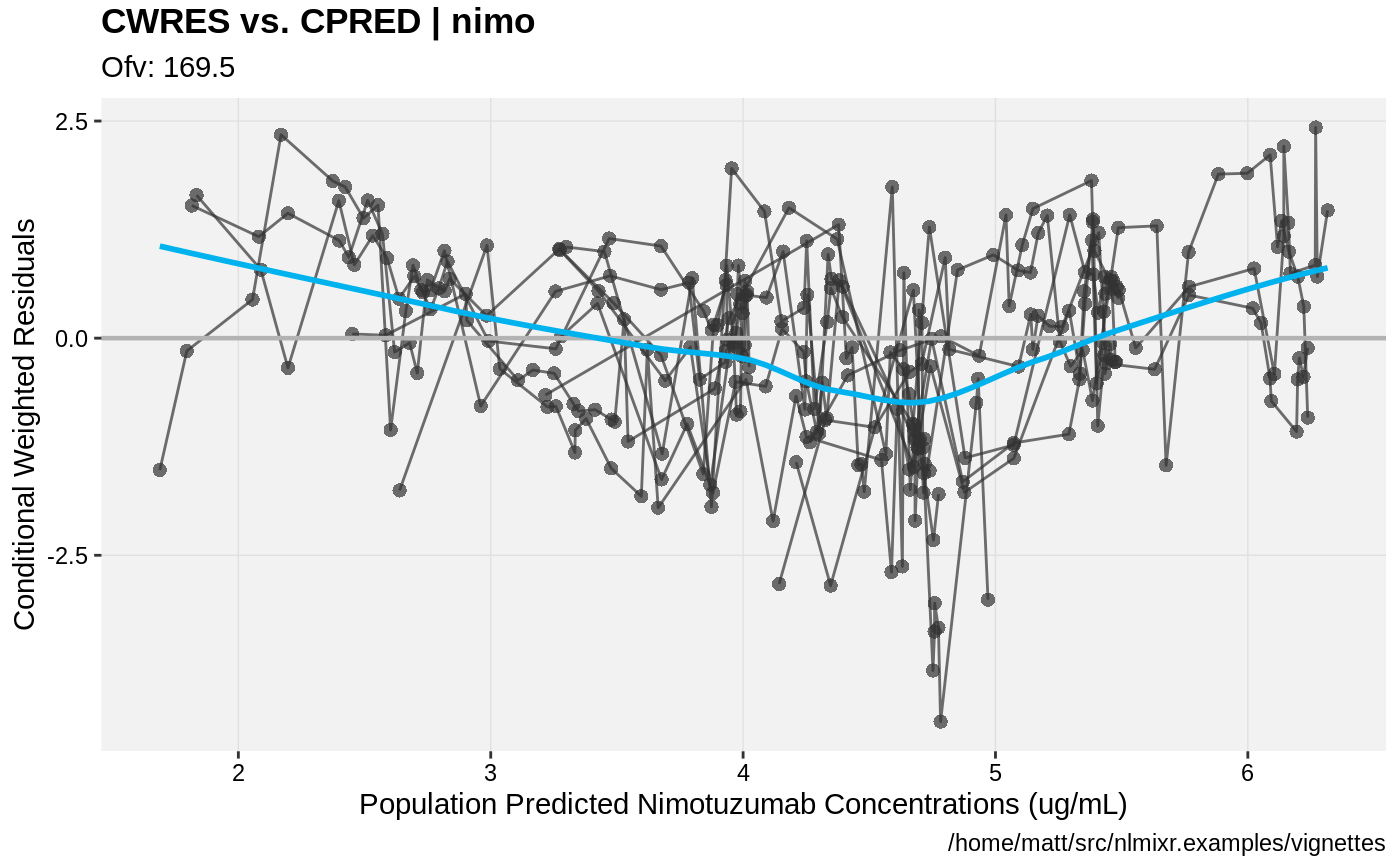
print(res_vs_idv(xpdb) +
ylab("Conditional Weighted Residuals") +
xlab("Time (h)"))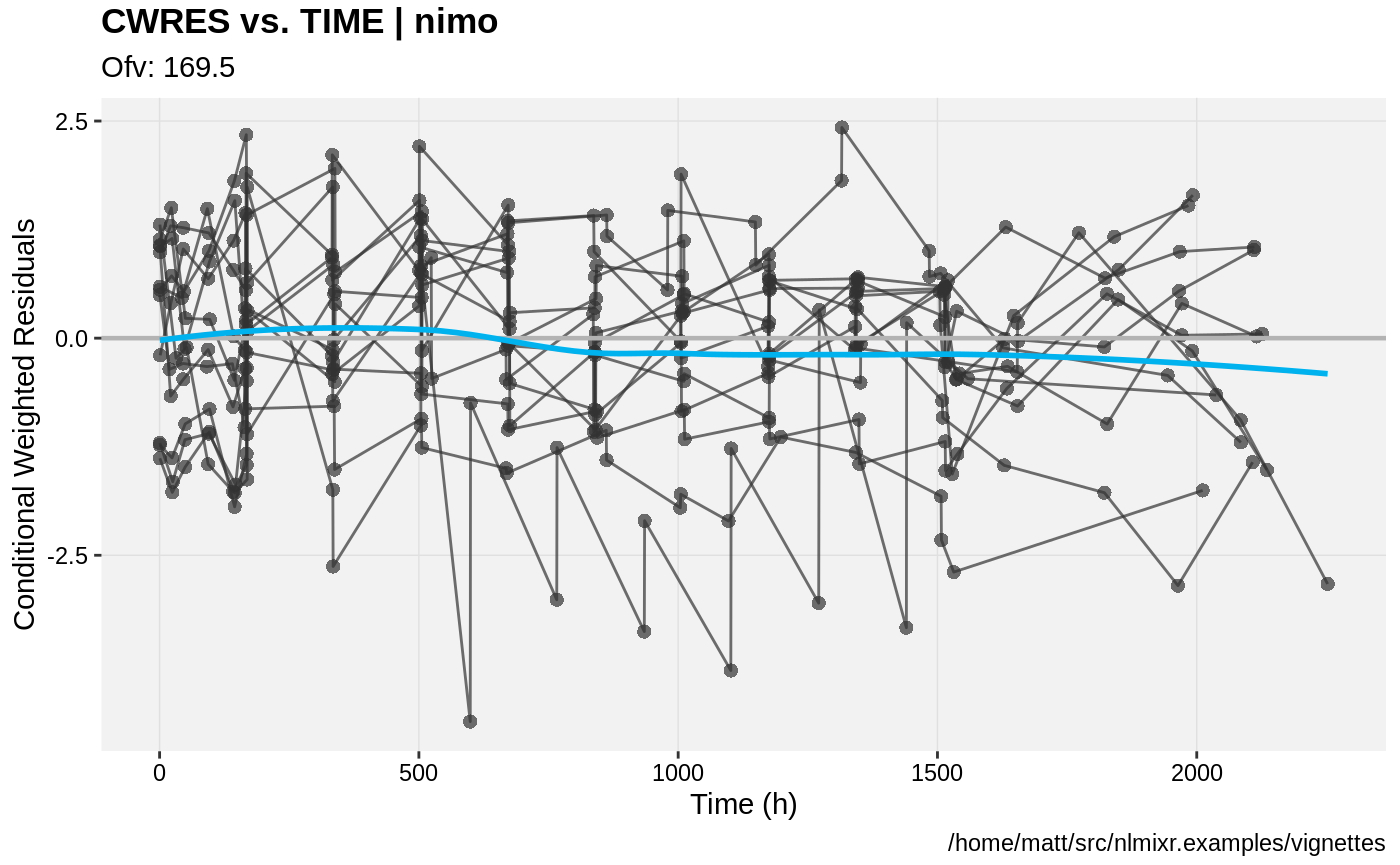
print(prm_vs_iteration(xpdb))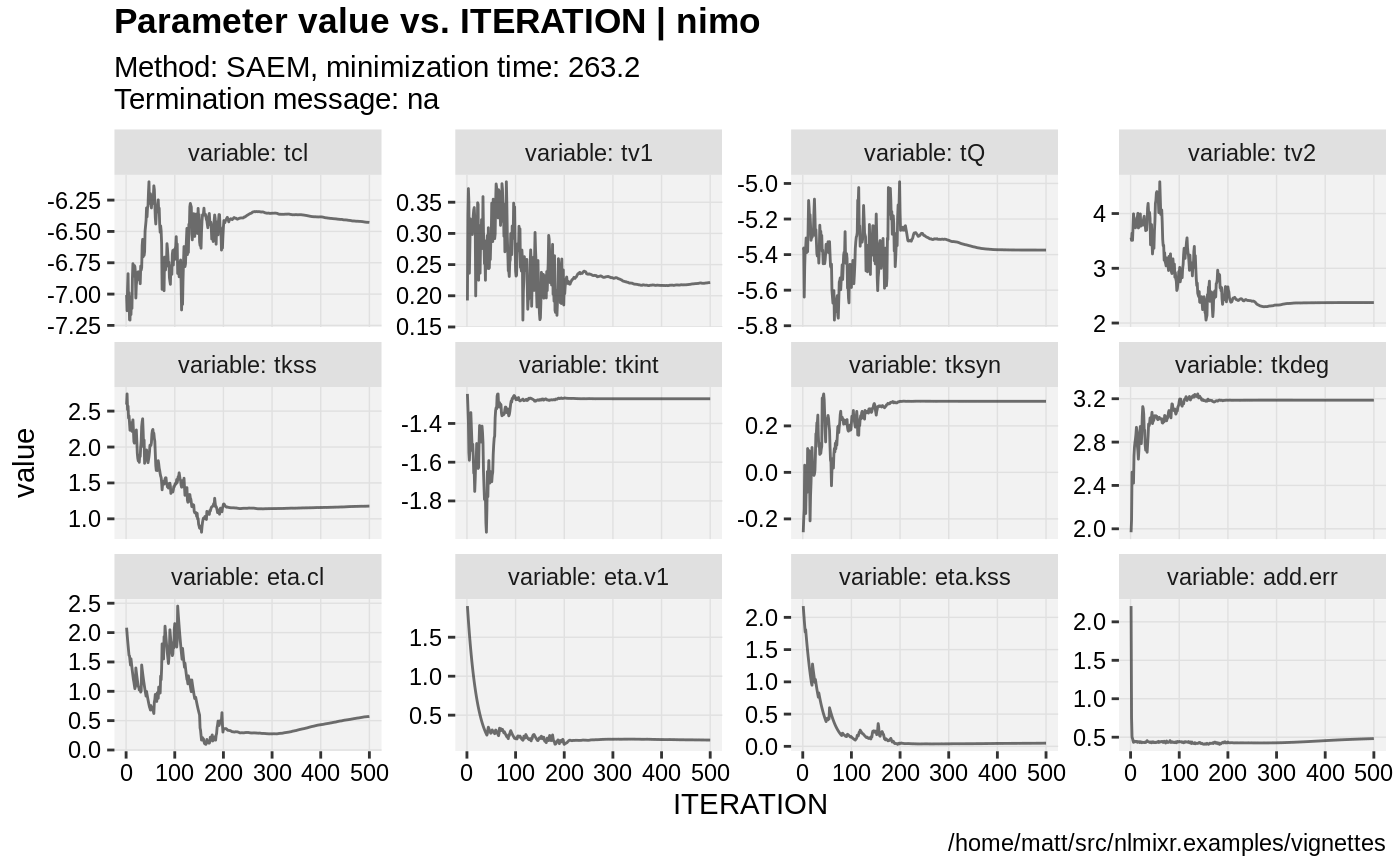
print(absval_res_vs_idv(xpdb, res = 'IWRES') +
ylab("Individual Weighted Residuals") +
xlab("Time (h)"))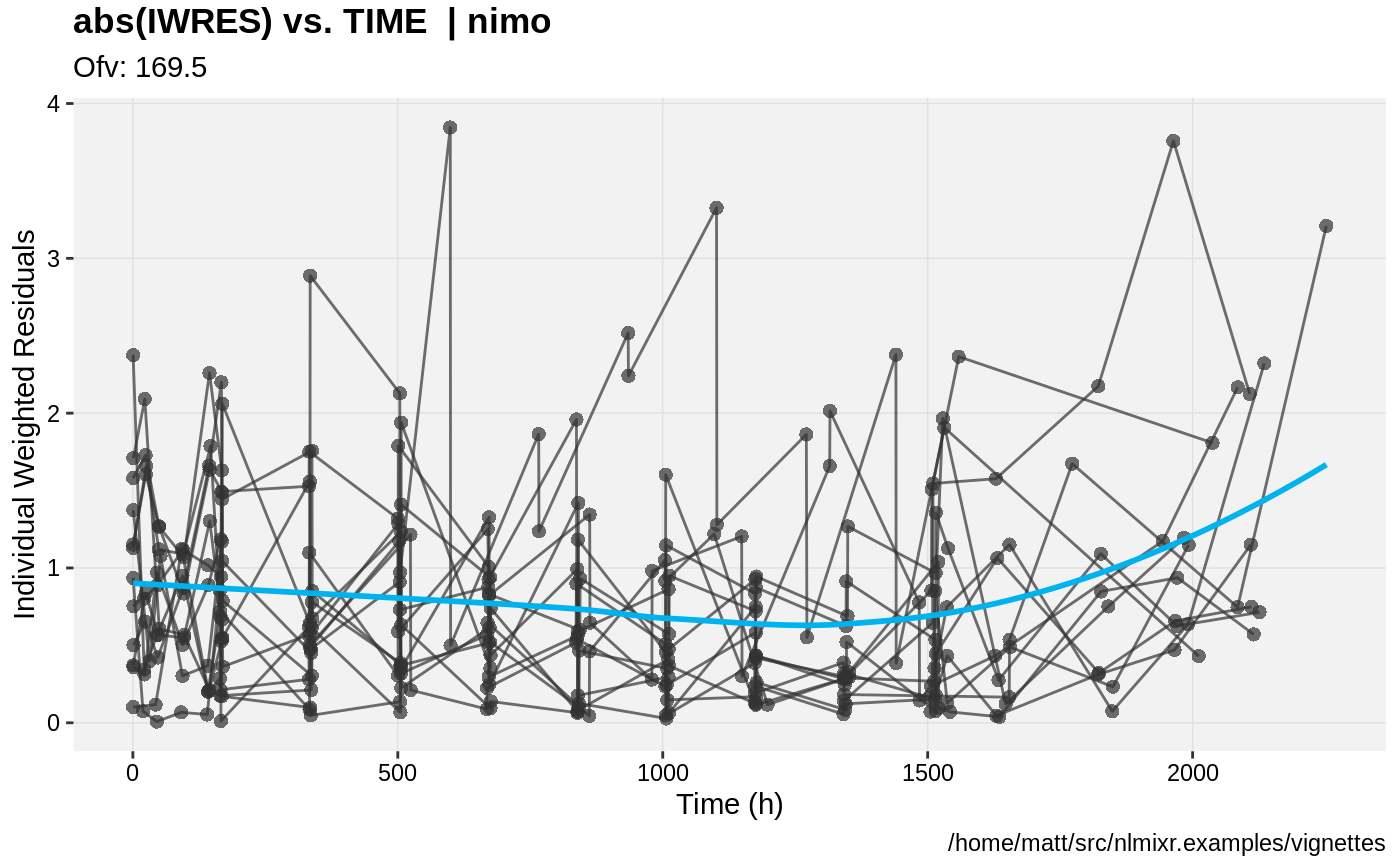
print(absval_res_vs_pred(xpdb, res = 'IWRES') +
ylab("Individual Weighted Residuals") +
xlab("Population Predicted Nimotuzumab Concentrations (ug/mL)"))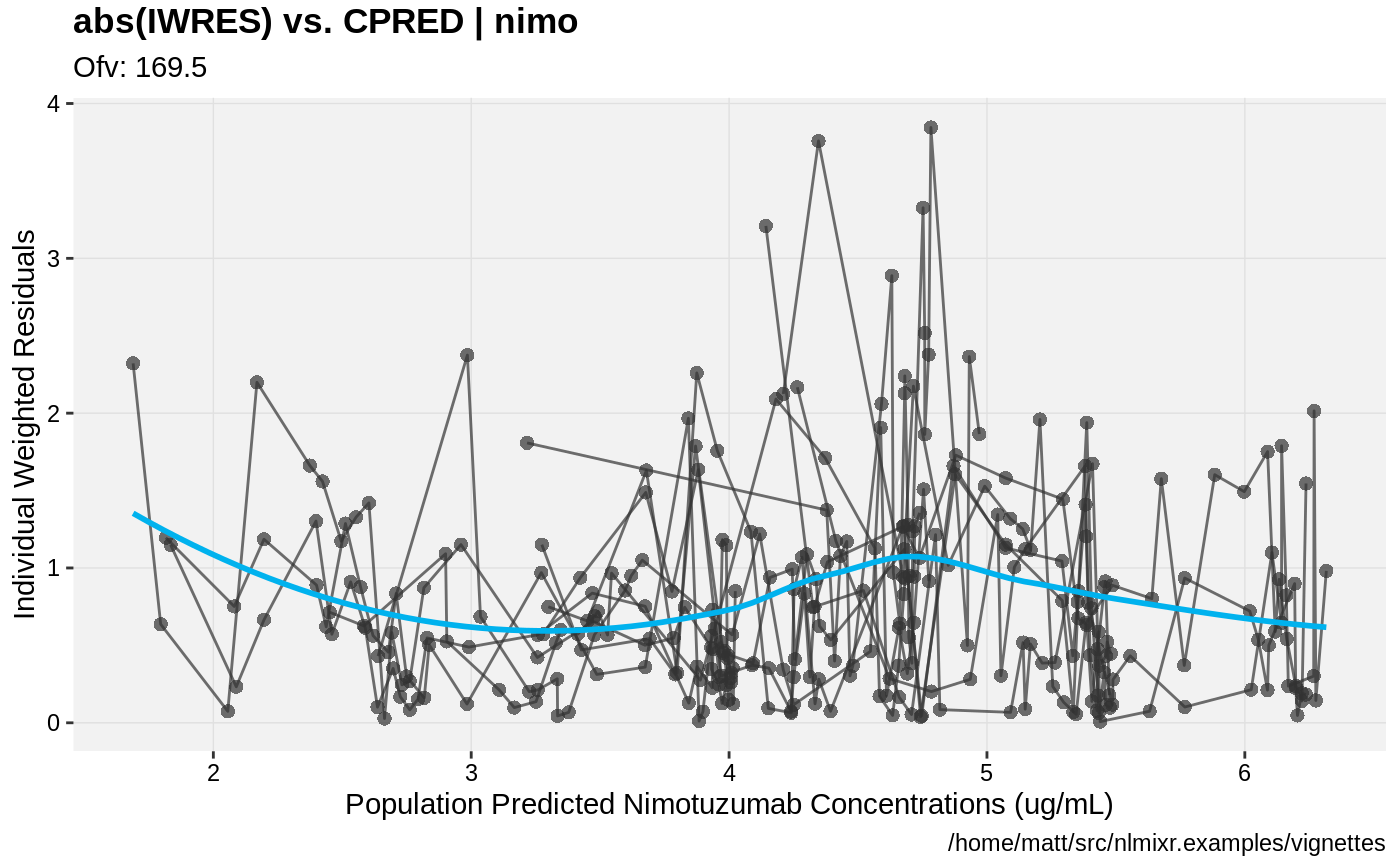
print(ind_plots(xpdb, nrow=3, ncol=4) +
ylab("Predicted and Observed Nimotuzumab concentrations (ug/mL)") +
xlab("Time (h)"))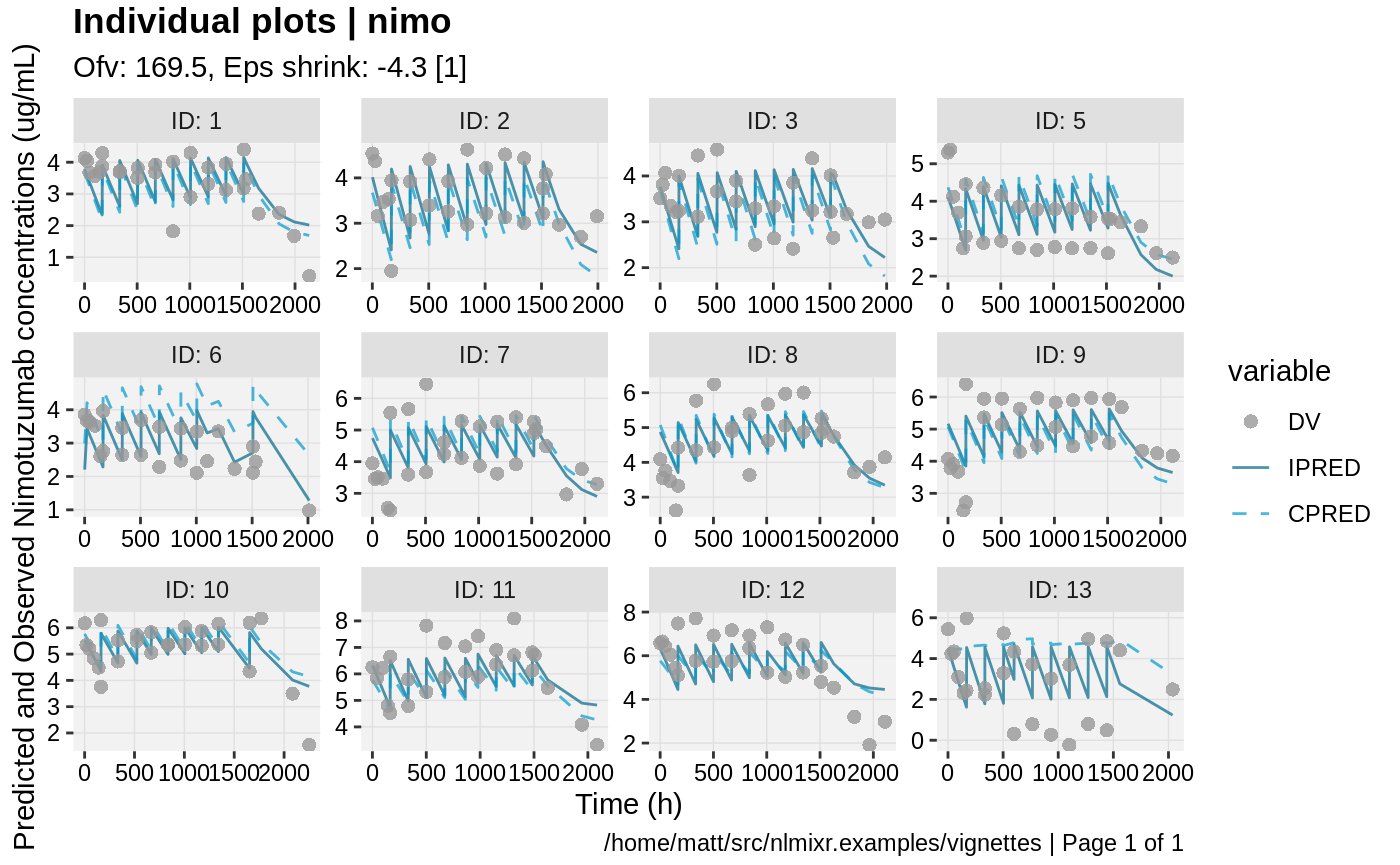
print(res_distrib(xpdb) +
ylab("Density") +
xlab("Conditional Weighted Residuals"))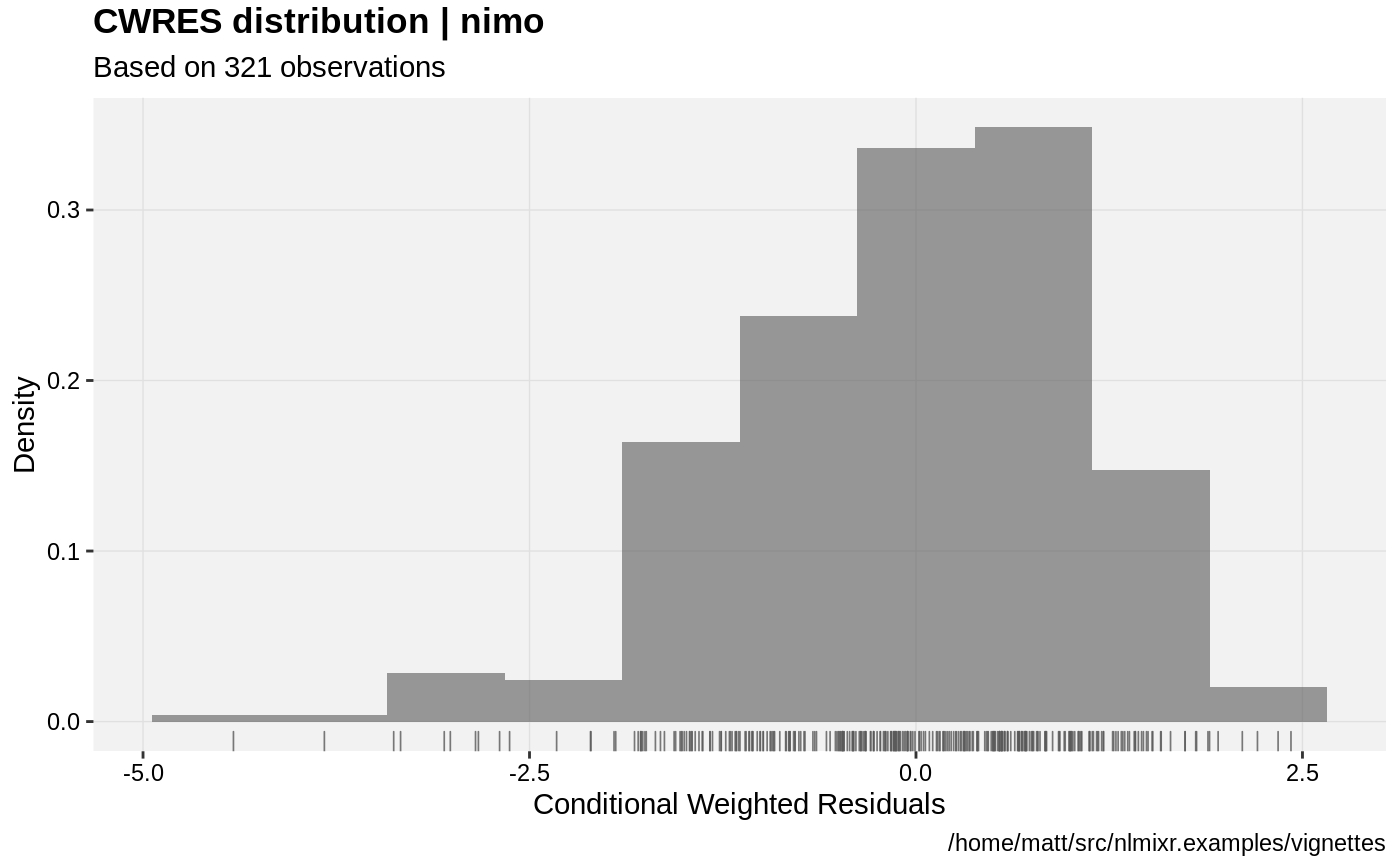
################################################################################
##Visual Predictive Checks
################################################################################
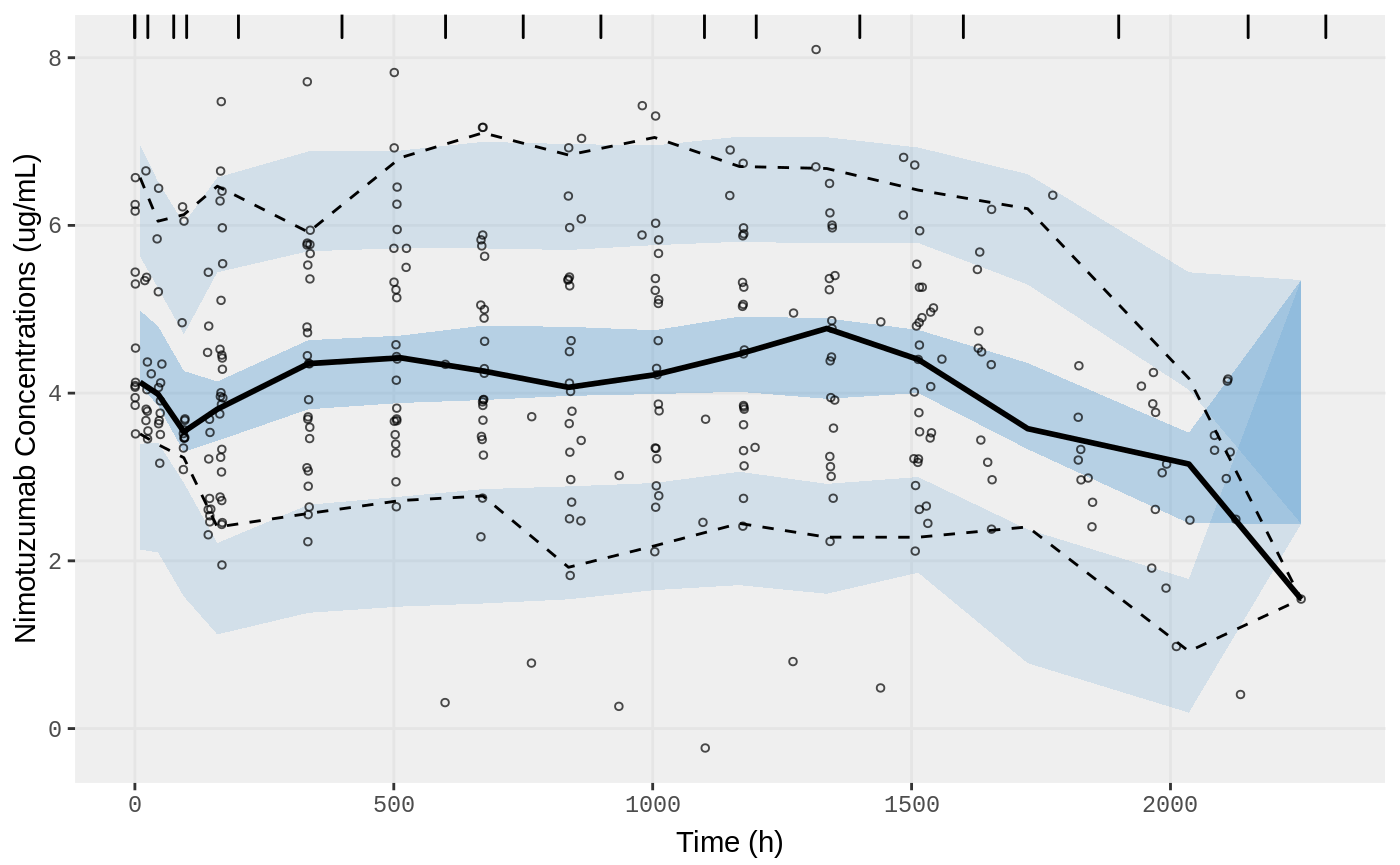
vpc.ui(fit,n=500,stratify=c("dos"), show=list(obs_dv=T),
bins = c(-0.5,0,25,75,100,200,400,600,750,900,1100,1200,1400,1600,1900,2150,2300),
ylab = "Nimotuzumab Concentrations (ug/mL)", xlab = "Time (h)")
#> Loading vpc already run (/tmp/RtmpXKdUFj/temp_libpath58be457a881a/nlmixr.examples/nlmixr-vpc-nimo-nimoData--f758cba02e5dd0415b091df7f94d896a.rds)
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now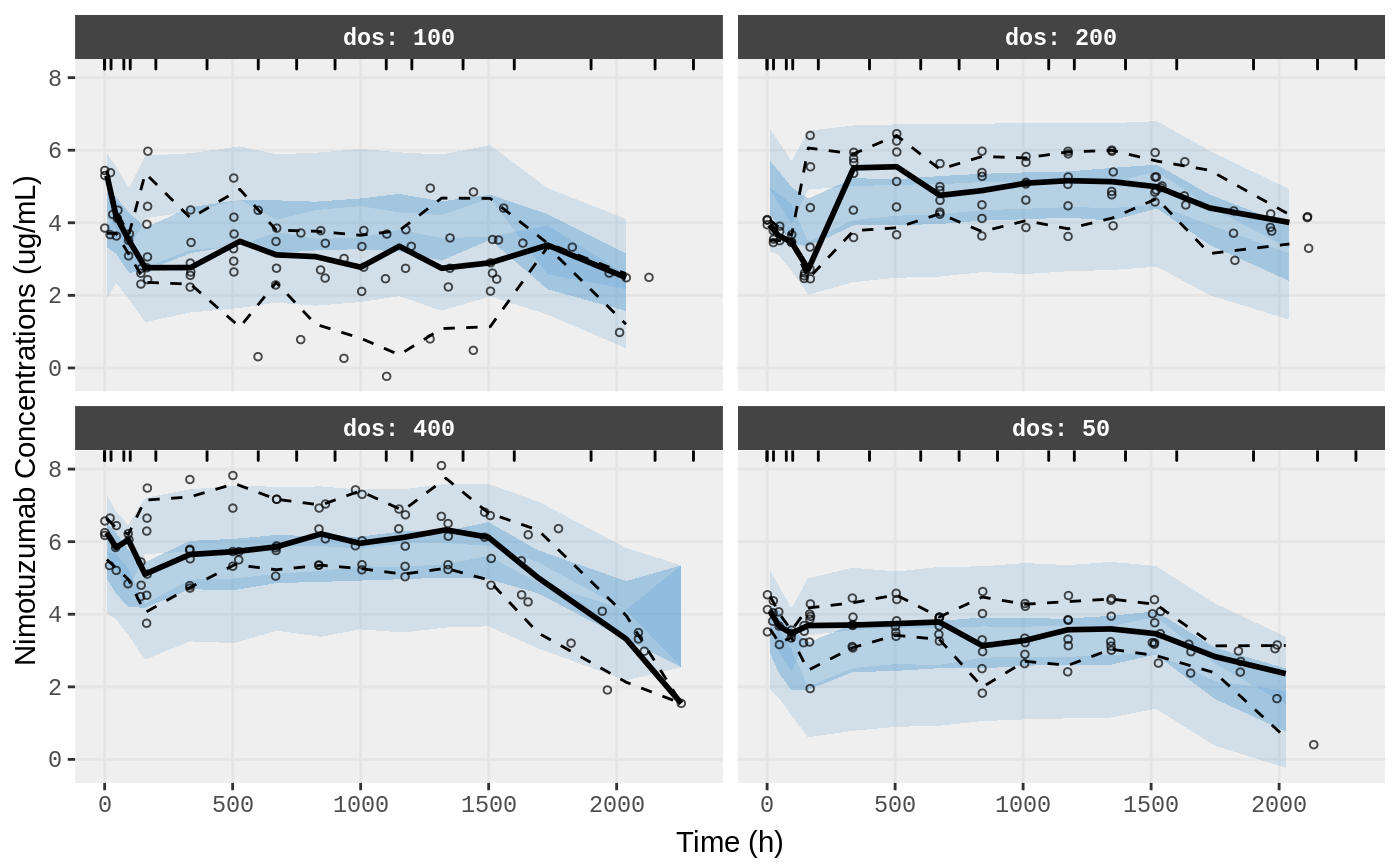
vpc.ui(fit,n=500, show=list(obs_dv=T),
bins = c(-0.5,0,25,75,100,200,400,600,750,900,1100,1200,1400,1600,1900,2150,2300),
ylab = "Nimotuzumab Concentrations (ug/mL)", xlab = "Time (h)")
#> Loading vpc already run (/tmp/RtmpXKdUFj/temp_libpath58be457a881a/nlmixr.examples/nlmixr-vpc-nimo-nimoData--8e579521023727c8f94f9200f08079d5.rds)
#> $rxsim = original simulated data
#> $sim = merge simulated data
#> $obs = observed data
#> $gg = vpc ggplot
#> use vpc(...) to change plot options
#> plotting the object now