Example Model: Phenobarbatol with correlations
2019-10-17
Source:vignettes/addingCovariances.Rmd
addingCovariances.Rmd
nlmixr
Adding Covariances between random effects
You can simply add co-variances between two random effects by adding the effects together in the model specification block, that is eta.cl+eta.v ~. After that statement, you specify the lower triangular matrix of the fit with c().
An example of this is the Phenobarbital data:
## Load Phenobarb data
library(nlmixr)Model Specification
pheno <- function() {
ini({
tcl <- log(0.008) # typical value of clearance
tv <- log(0.6) # typical value of volume
## var(eta.cl)
eta.cl + eta.v ~ c(1,
0.01, 1) ## cov(eta.cl, eta.v), var(eta.v)
# interindividual variability on clearance and volume
add.err <- 0.1 # residual variability
})
model({
cl <- exp(tcl + eta.cl) # individual value of clearance
v <- exp(tv + eta.v) # individual value of volume
ke <- cl / v # elimination rate constant
d/dt(A1) = - ke * A1 # model differential equation
cp = A1 / v # concentration in plasma
cp ~ add(add.err) # define error model
})
}Fit with SAEM
fit <- nlmixr(pheno, pheno_sd, "saem", table=list(cwres=TRUE, npde=TRUE))
print(fit)
#> ── nlmixr SAEM([3mODE[23m); [2m[3mOBJF by FOCEi approximation[23m[22m fit ──────────────────────
#> OBJF AIC BIC Log-likelihood Condition Number
#> FOCEi 688.6326 985.5036 1003.764 -486.7518 7.536808
#>
#> ── Time (sec; $time): ─────────────────────────────────────────────────────
#> saem setup table cwres covariance npde other
#> elapsed -31.188 58.27651 0.011 58.296 0.019 0.604 0.918492
#>
#> ── Population Parameters ($parFixed or $parFixedDf): ──────────────────────
#> Parameter Est. SE %RSE
#> tcl typical value of clearance -5 0.0752 1.5
#> tv typical value of volume 0.346 0.0537 15.5
#> add.err residual variability 2.83
#> Back-transformed(95%CI) BSV(CV%) Shrink(SD)%
#> tcl 0.00677 (0.00584, 0.00784) 53.1 1.86%
#> tv 1.41 (1.27, 1.57) 40.9 1.25%
#> add.err 2.83
#>
#> Covariance Type ($covMethod): linFim
#> Correlations in between subject variability (BSV) matrix:
#> cor__eta.v.eta.cl
#> 0.985
#>
#> Full BSV covariance ($omega) or correlation ($omegaR; diagonals=SDs)
#> Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
#>
#> ── Fit Data (object is a modified tibble): ────────────────────────────────
#> # A tibble: 155 x 23
#> ID TIME DV EVID PRED RES WRES IPRED IRES IWRES CPRED
#> <fct> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 2 17.3 0 17.5 -0.228 -0.105 18.5 -1.16 -0.410 17.5
#> 2 1 112. 31 0 27.9 3.10 1.43 29.6 1.37 0.483 27.9
#> 3 2 2 9.7 0 10.5 -0.817 -0.376 12.5 -2.80 -0.989 10.3
#> # … with 152 more rows, and 12 more variables: CRES <dbl>, CWRES <dbl>,
#> # eta.cl <dbl>, eta.v <dbl>, cl <dbl>, v <dbl>, ke <dbl>, cp <dbl>,
#> # A1 <dbl>, EPRED <dbl>, ERES <dbl>, NPDE <dbl>Basic Goodness of Fit Plots
plot(fit)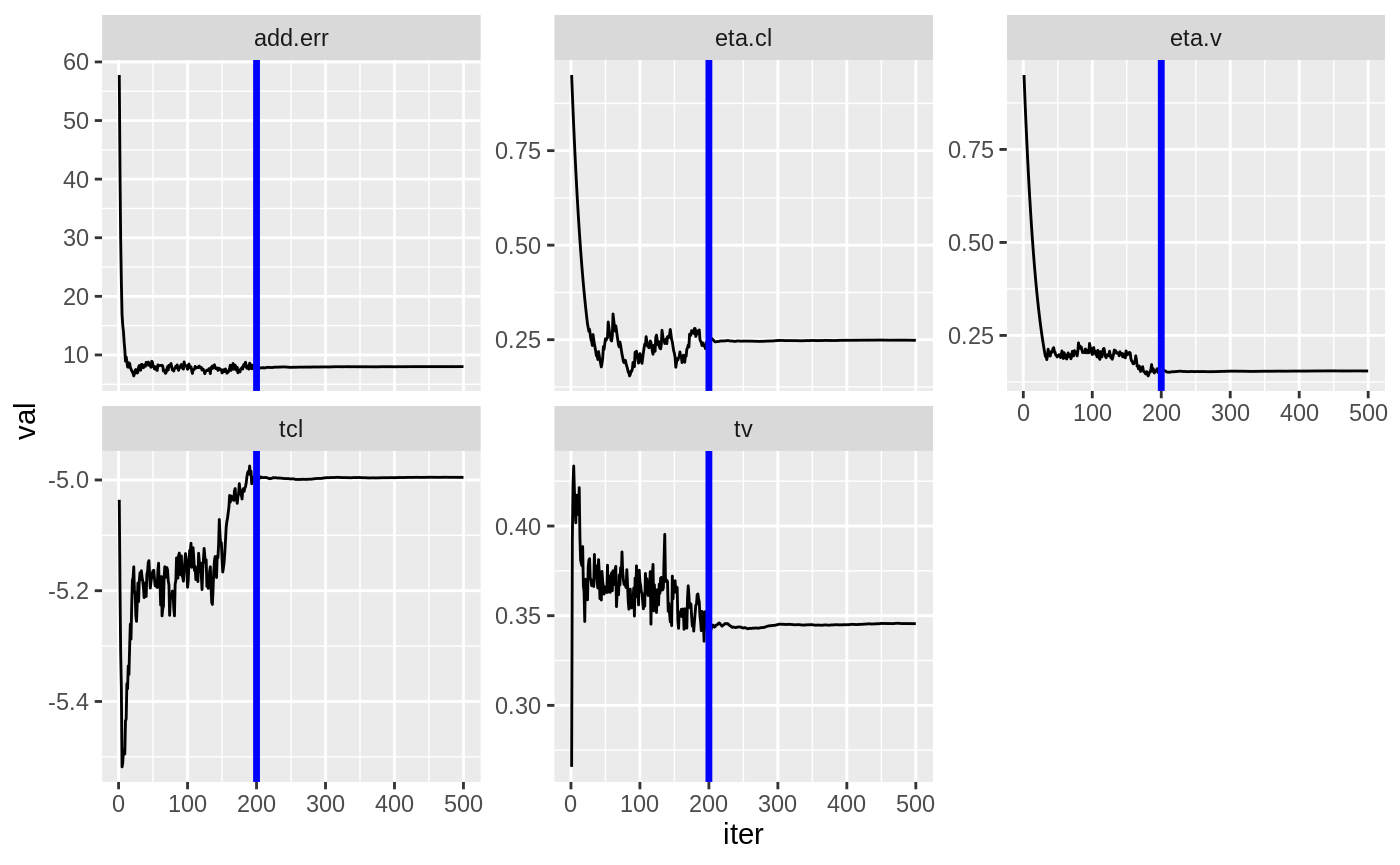
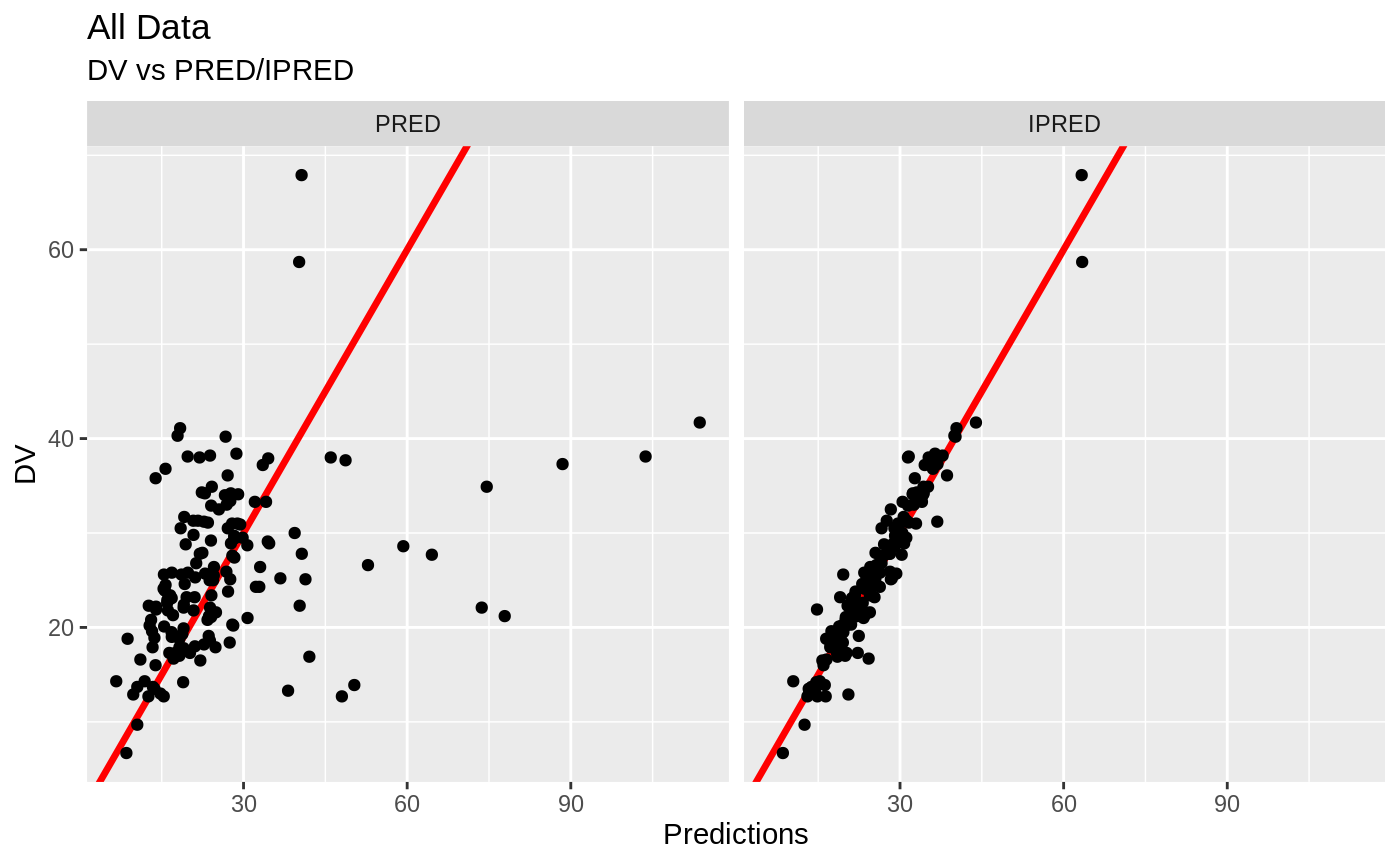
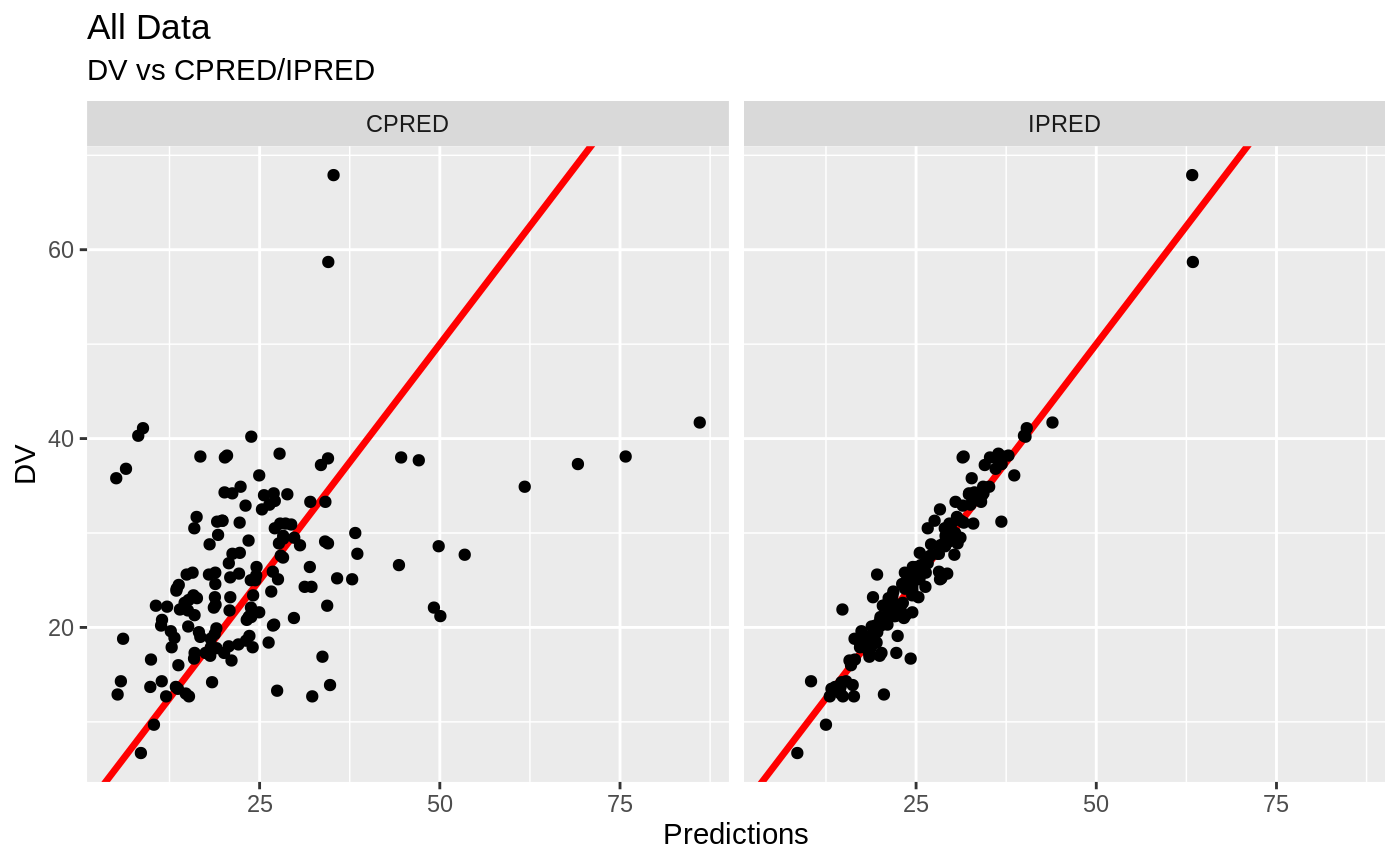
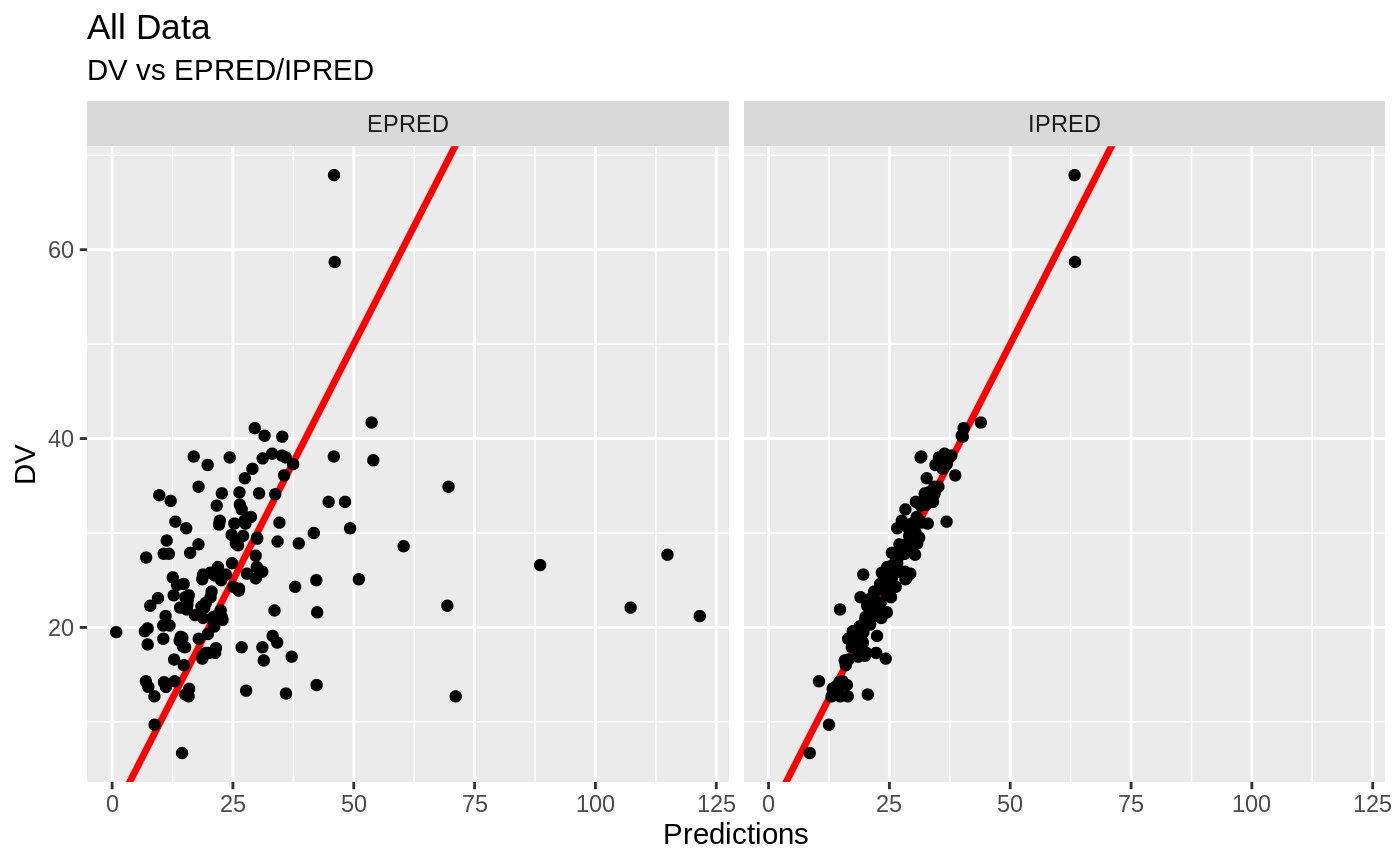
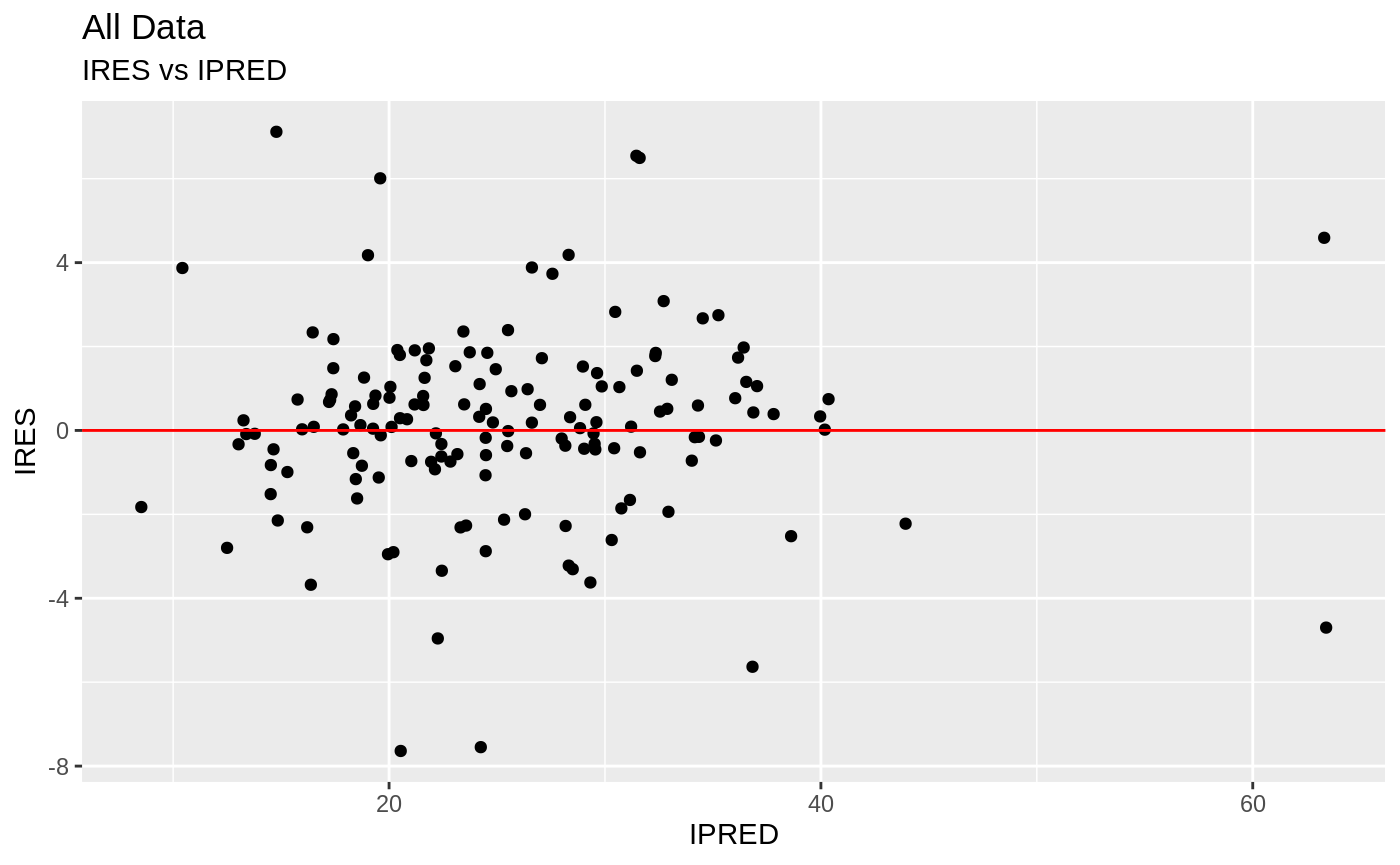
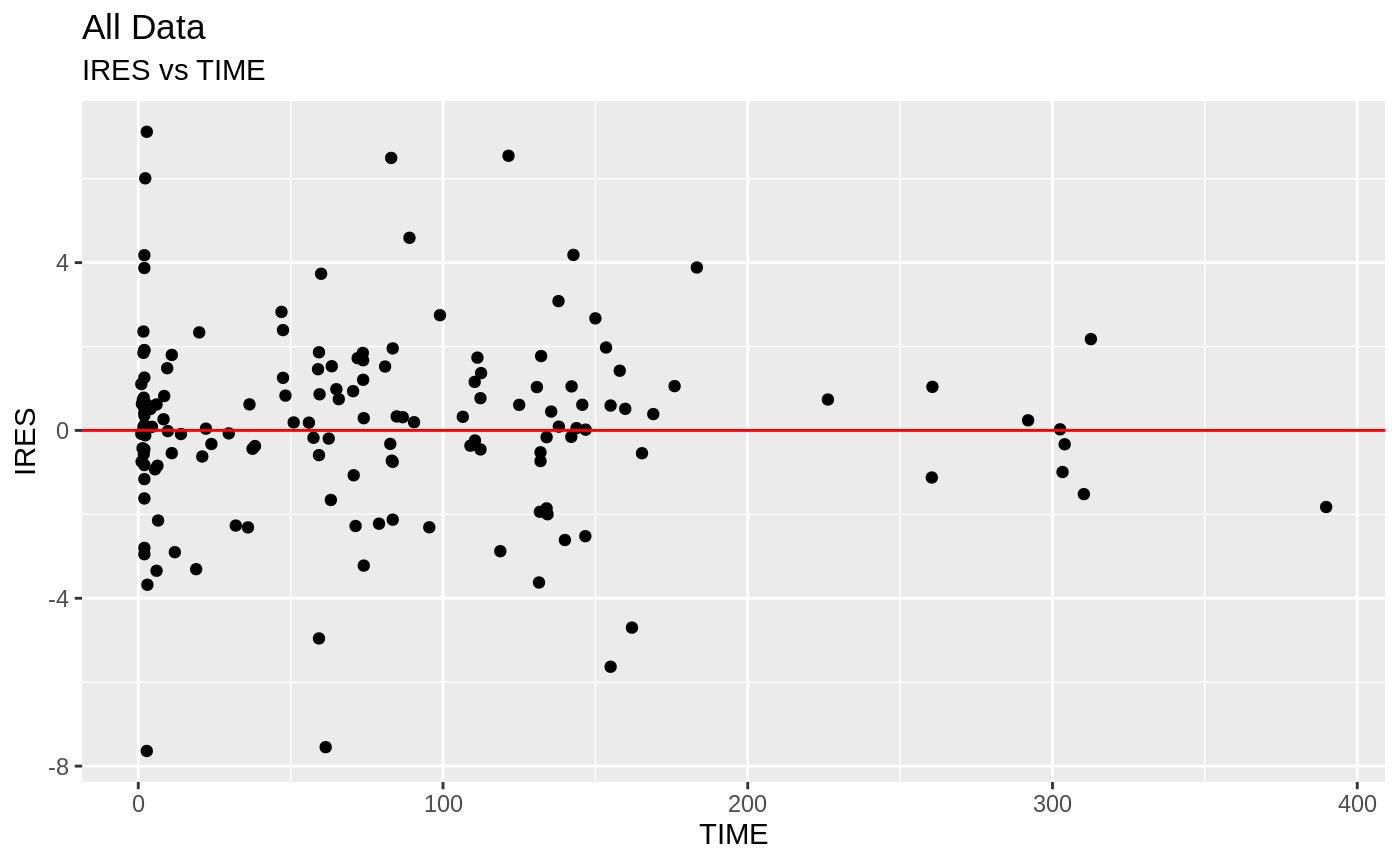
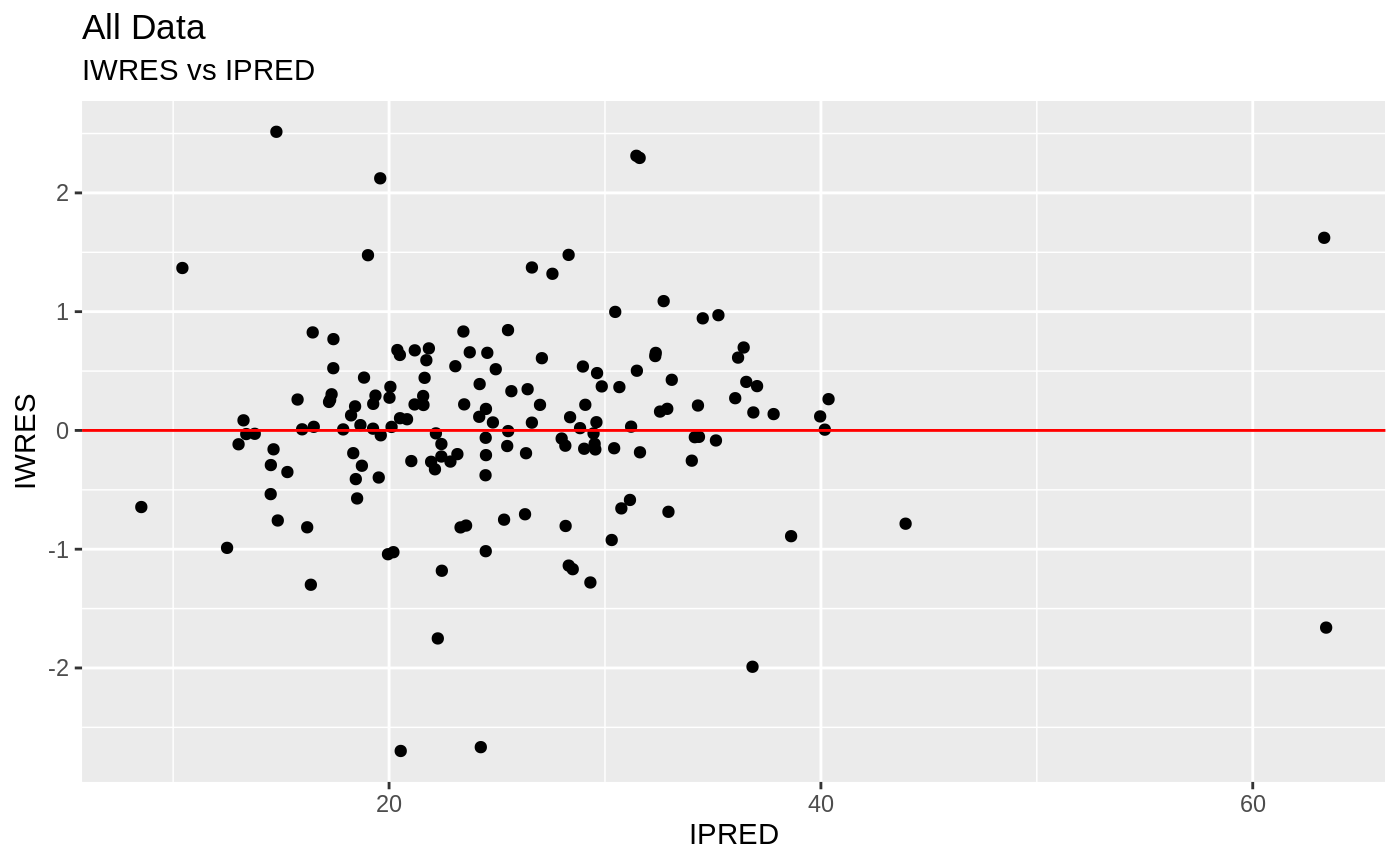
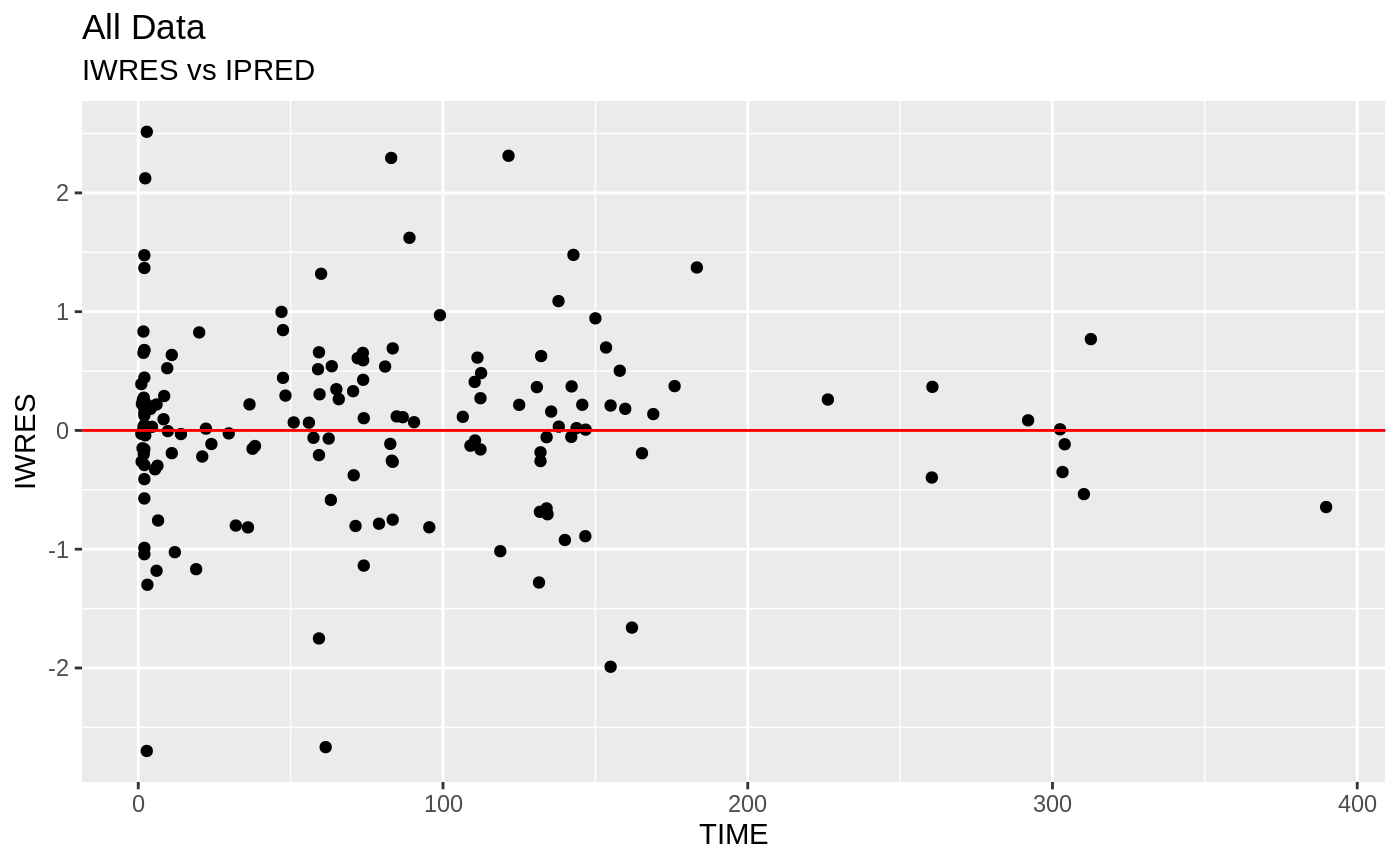
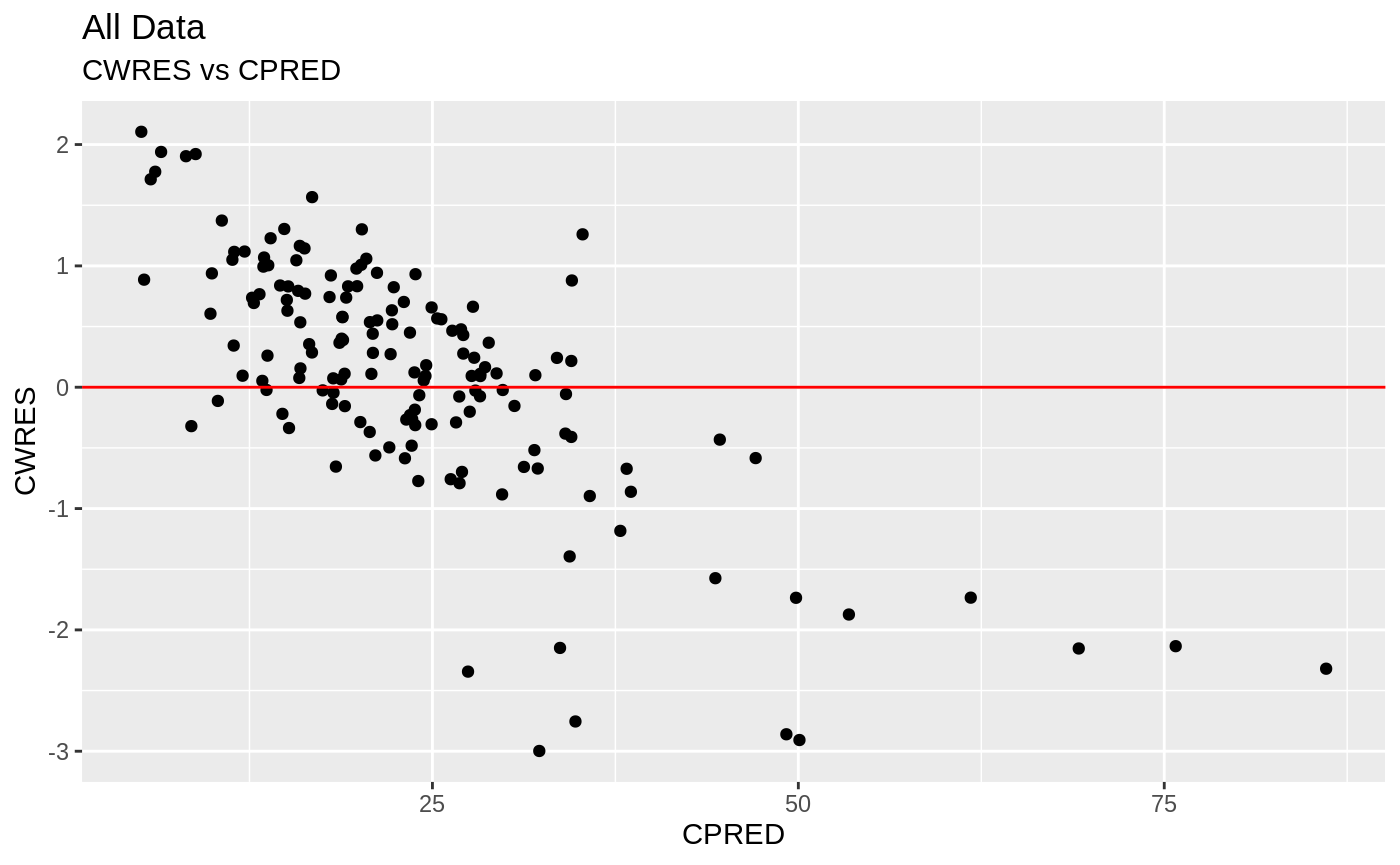
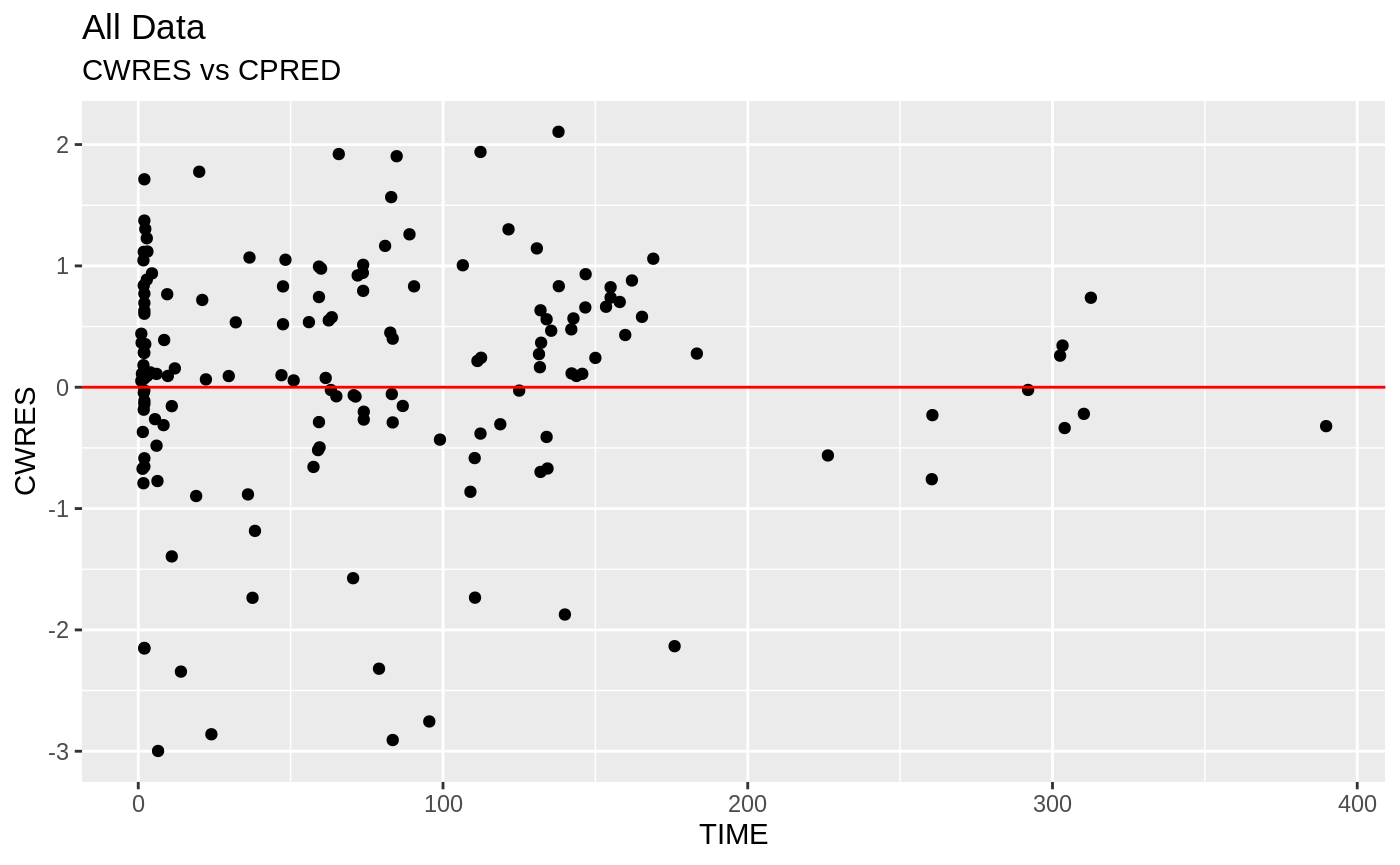
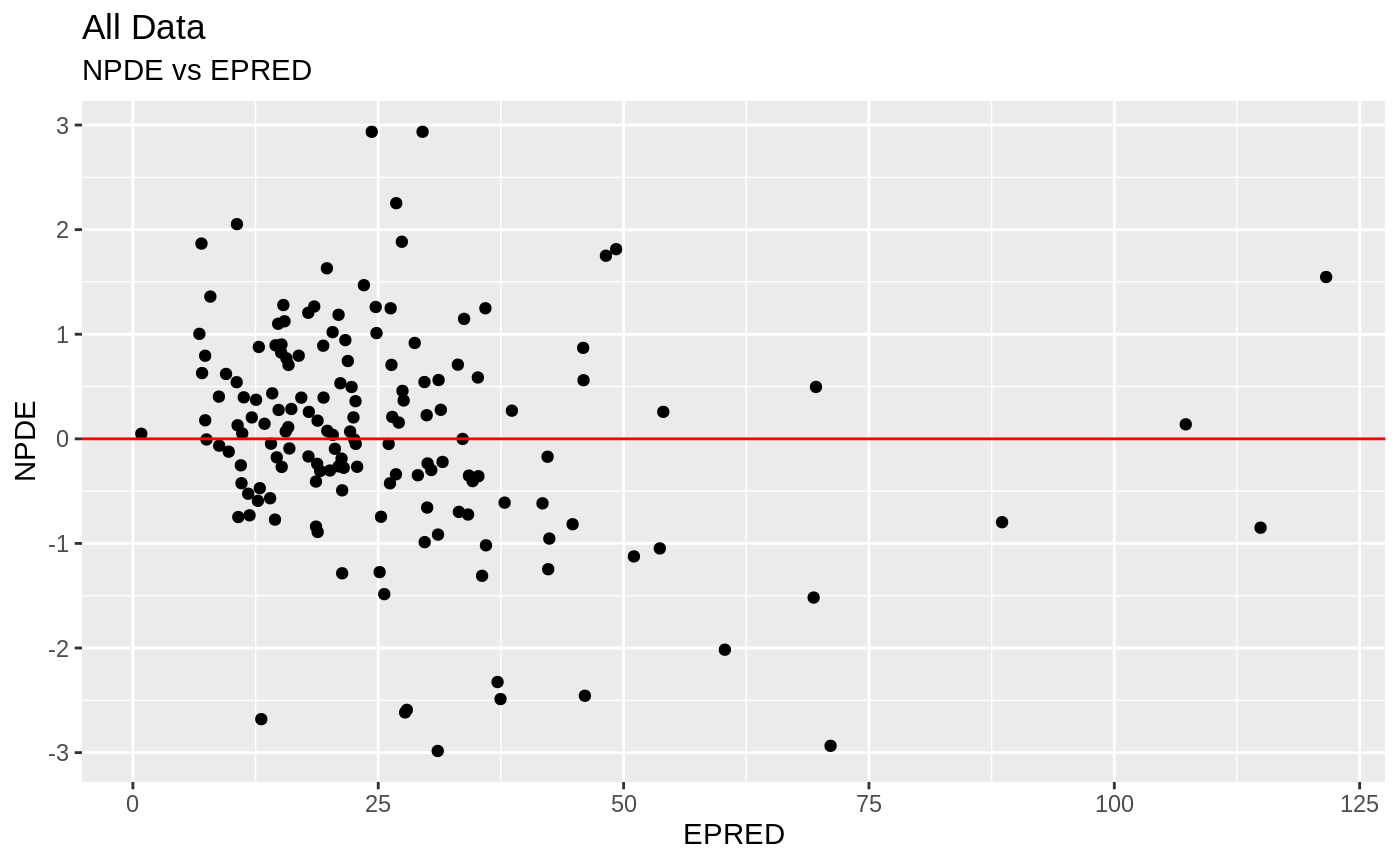
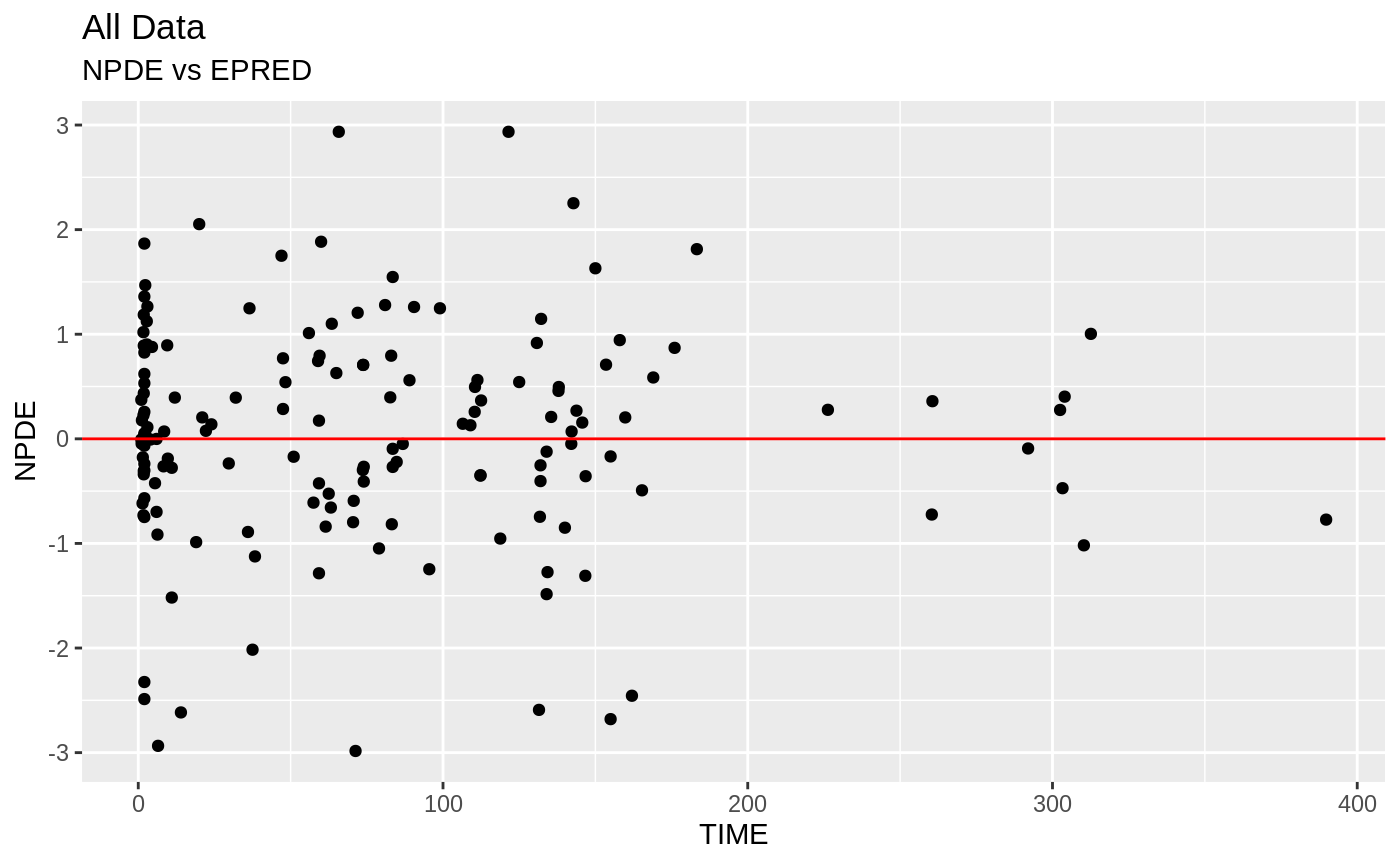
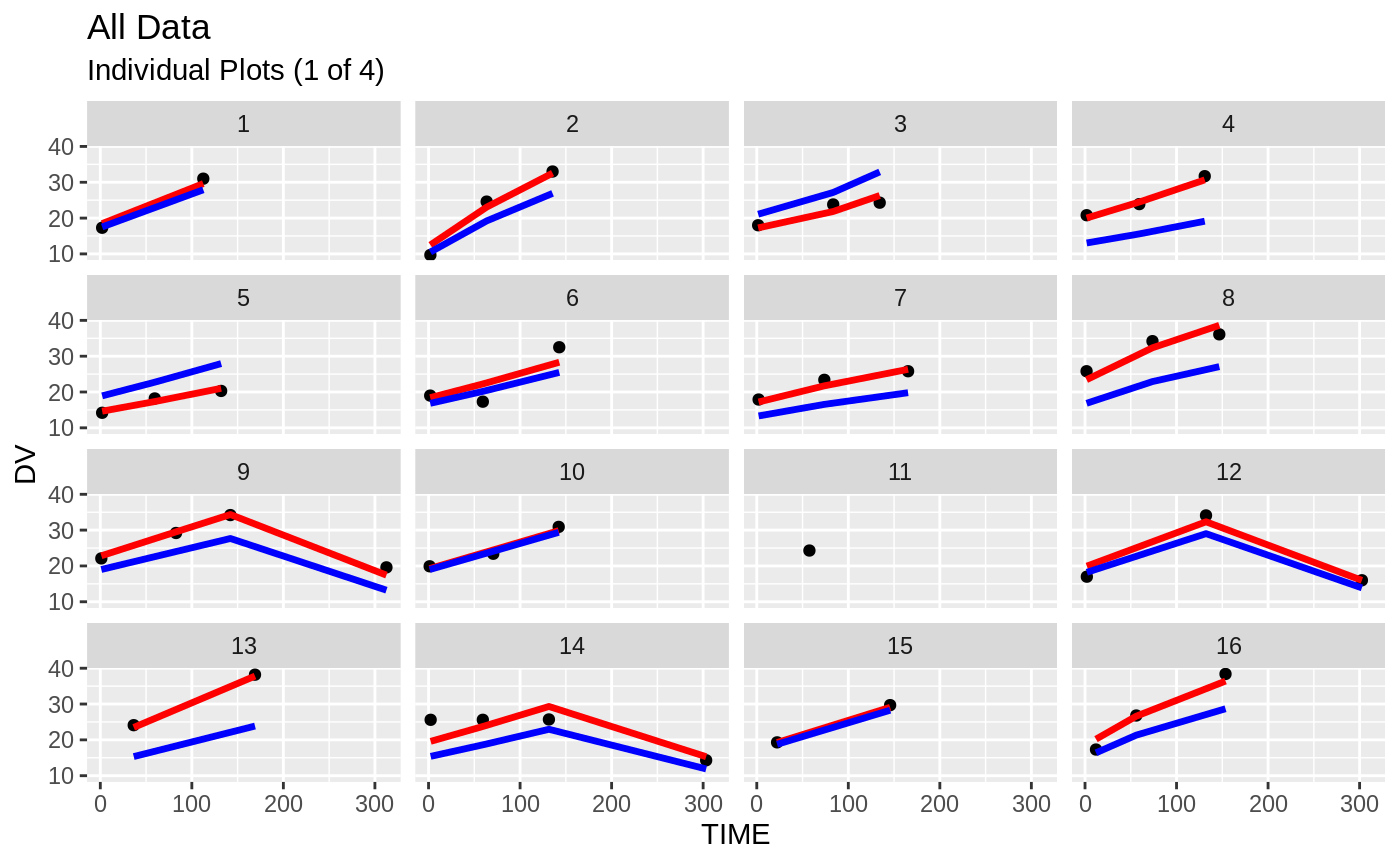
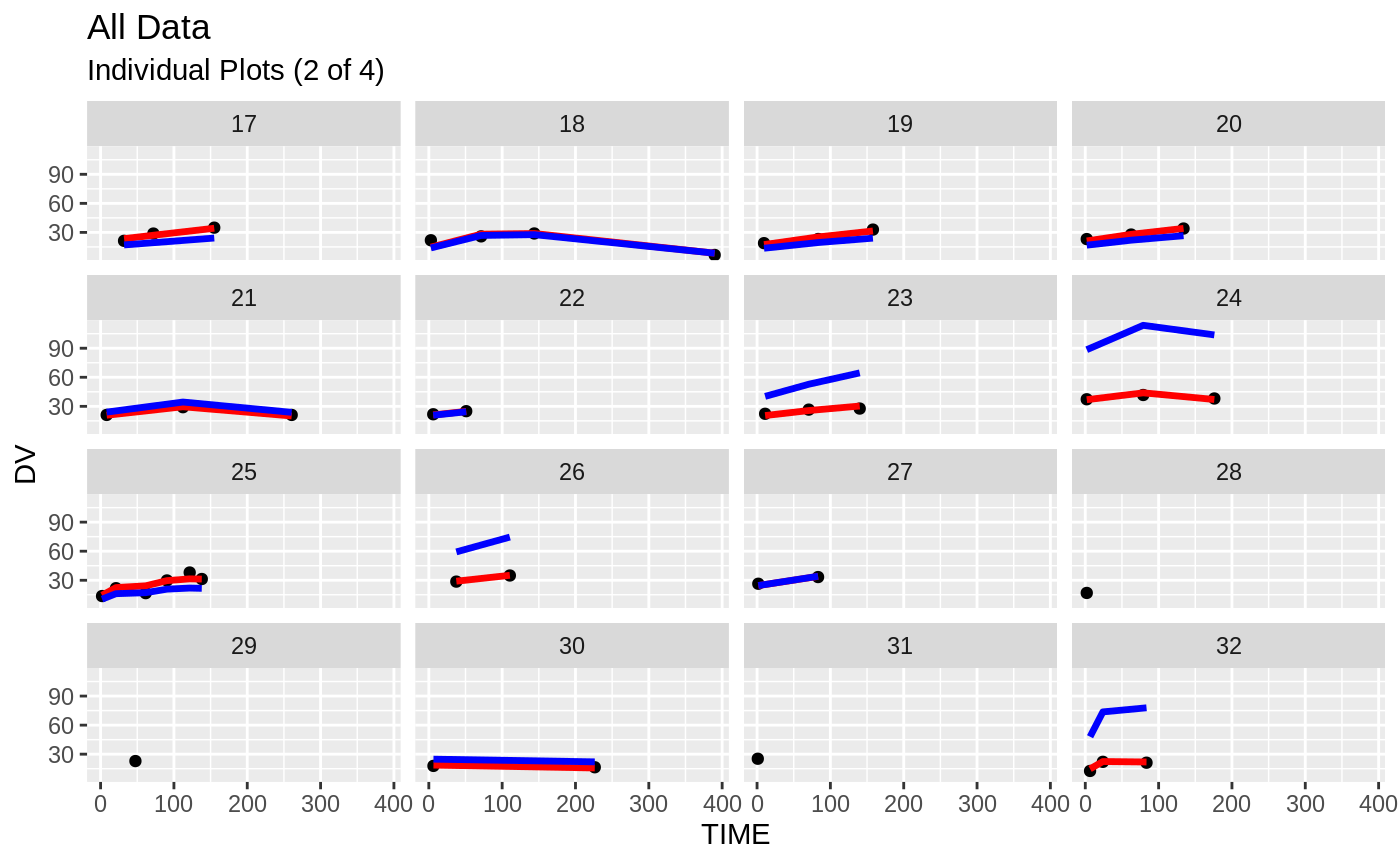
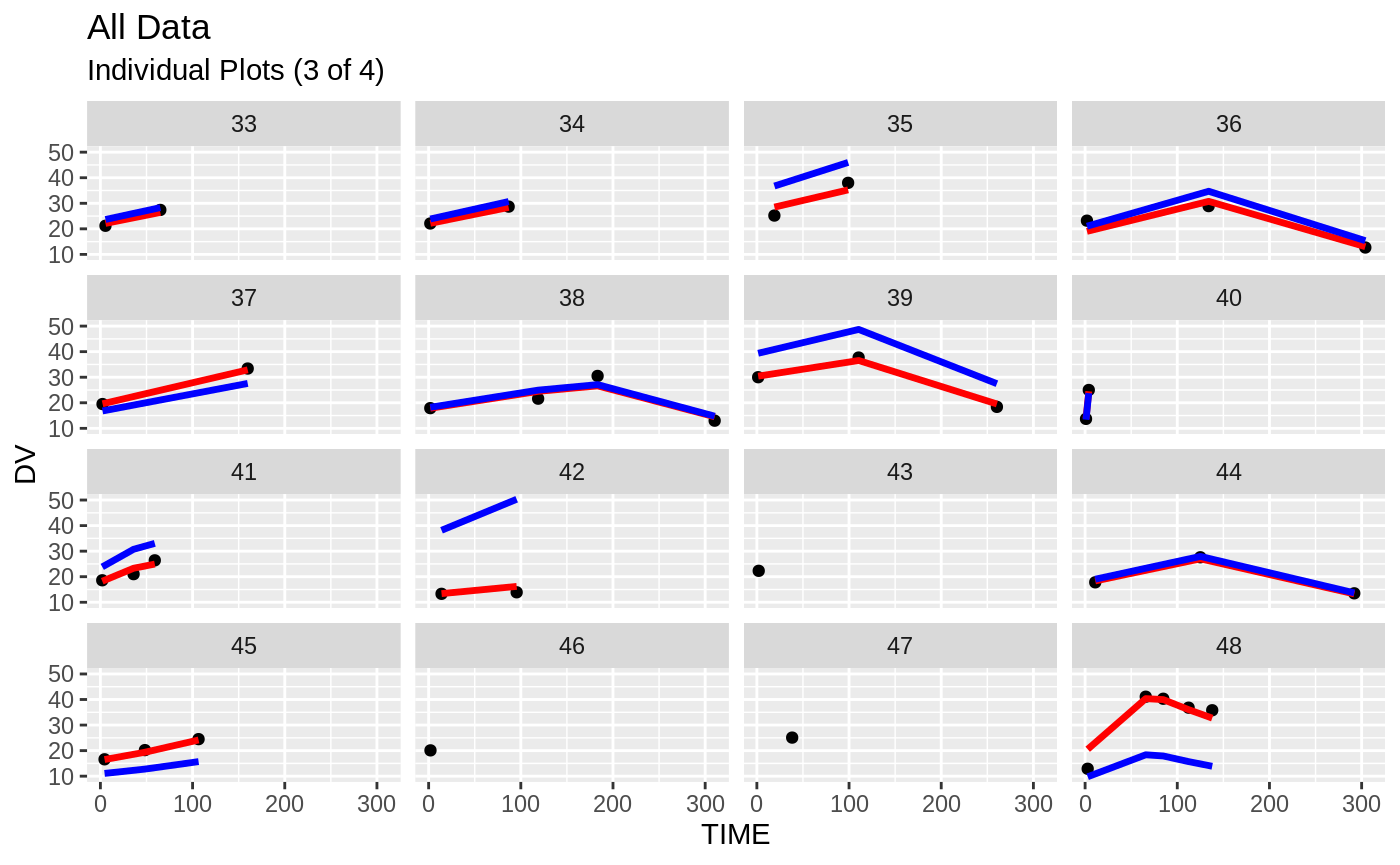
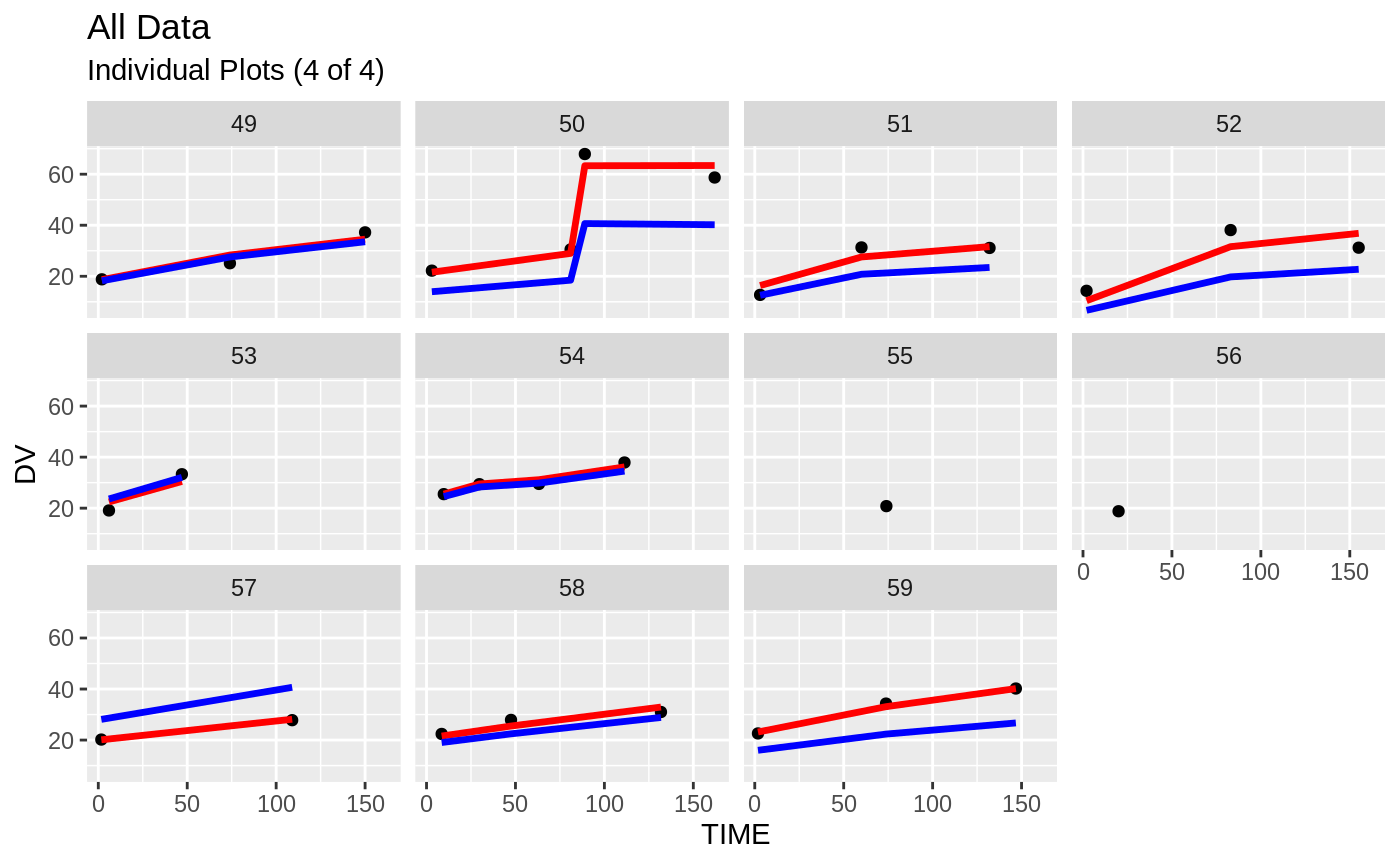
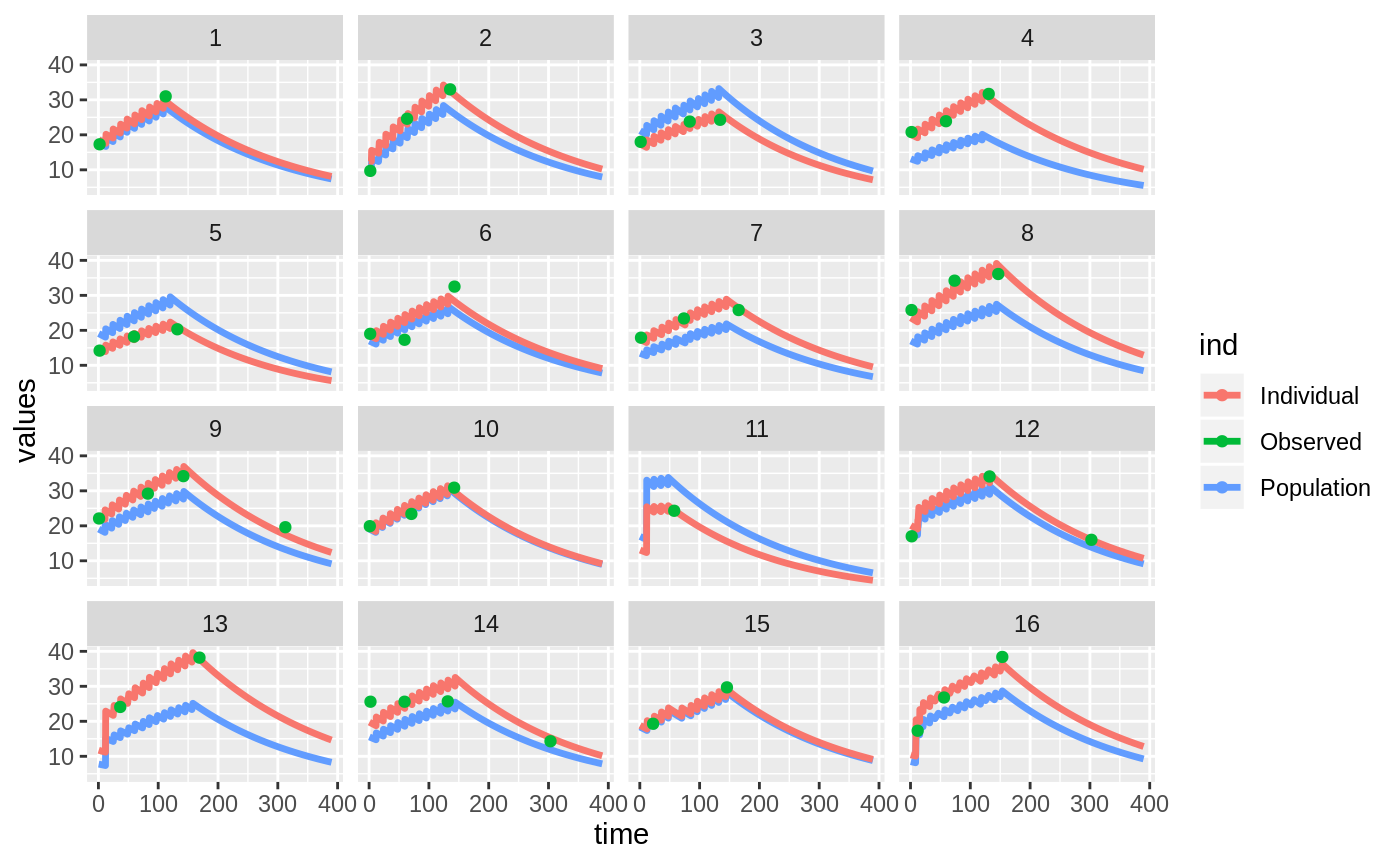
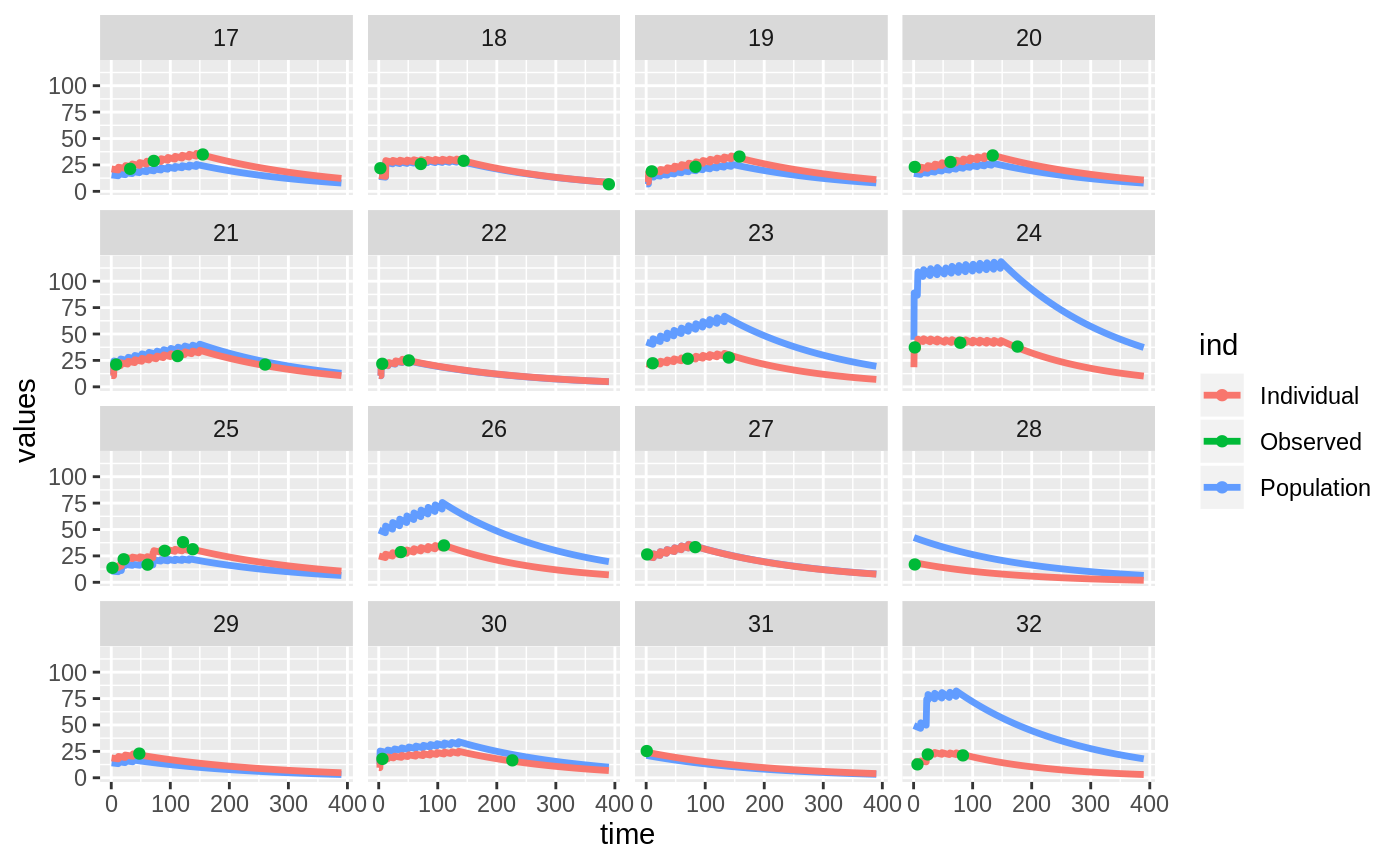
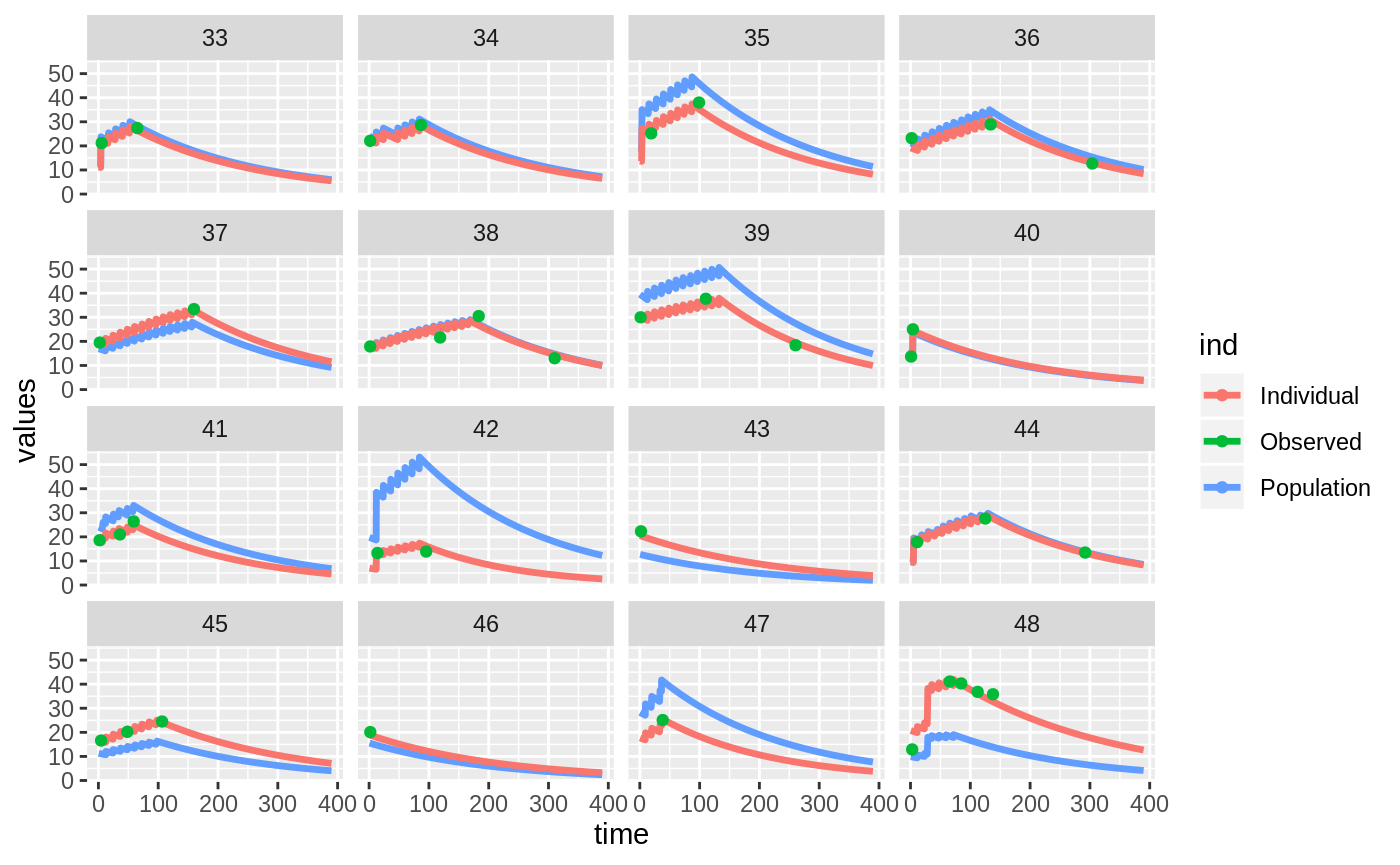
Those individual plots are not that great, it would be better to see the actual curves; You can with augPred
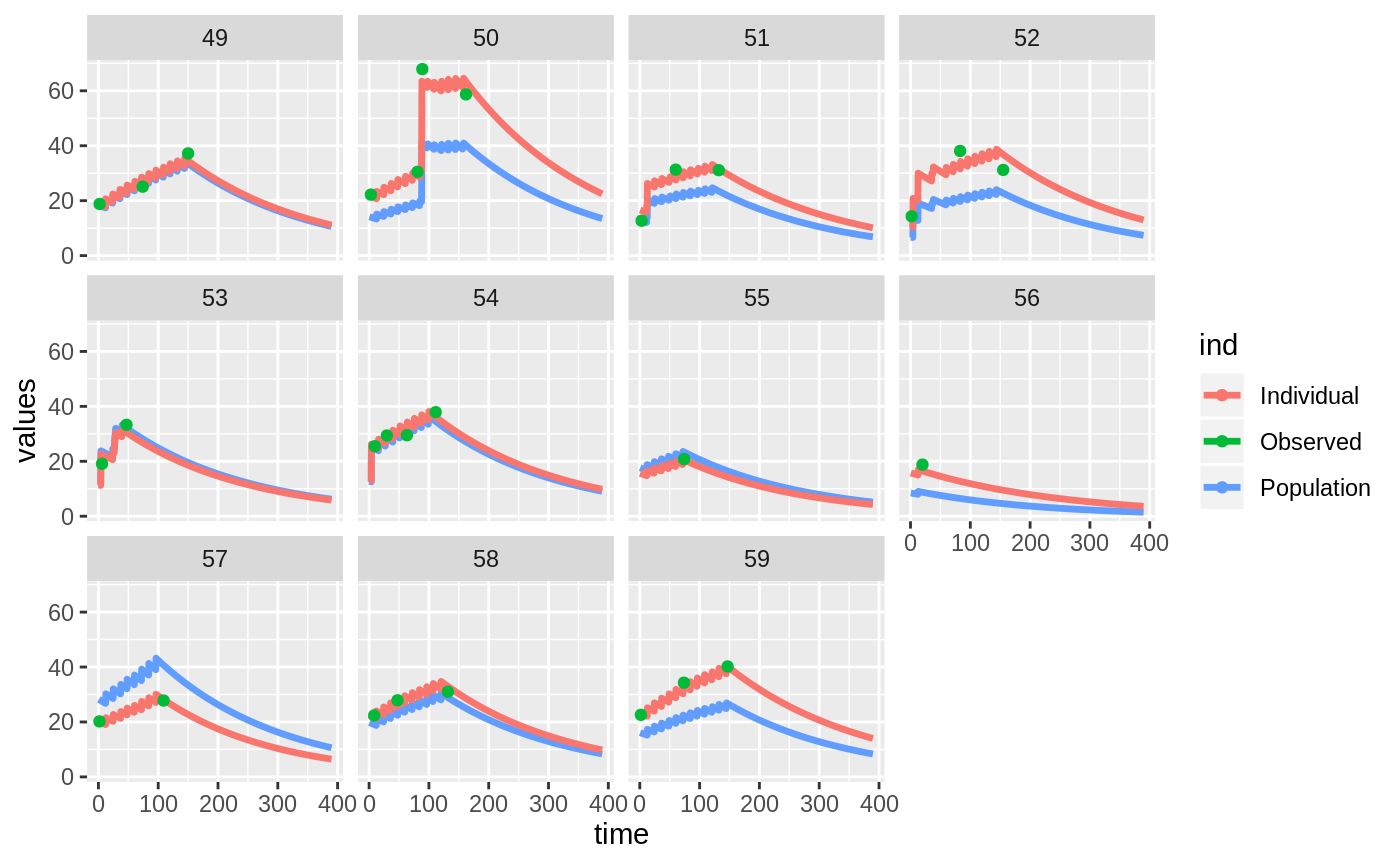
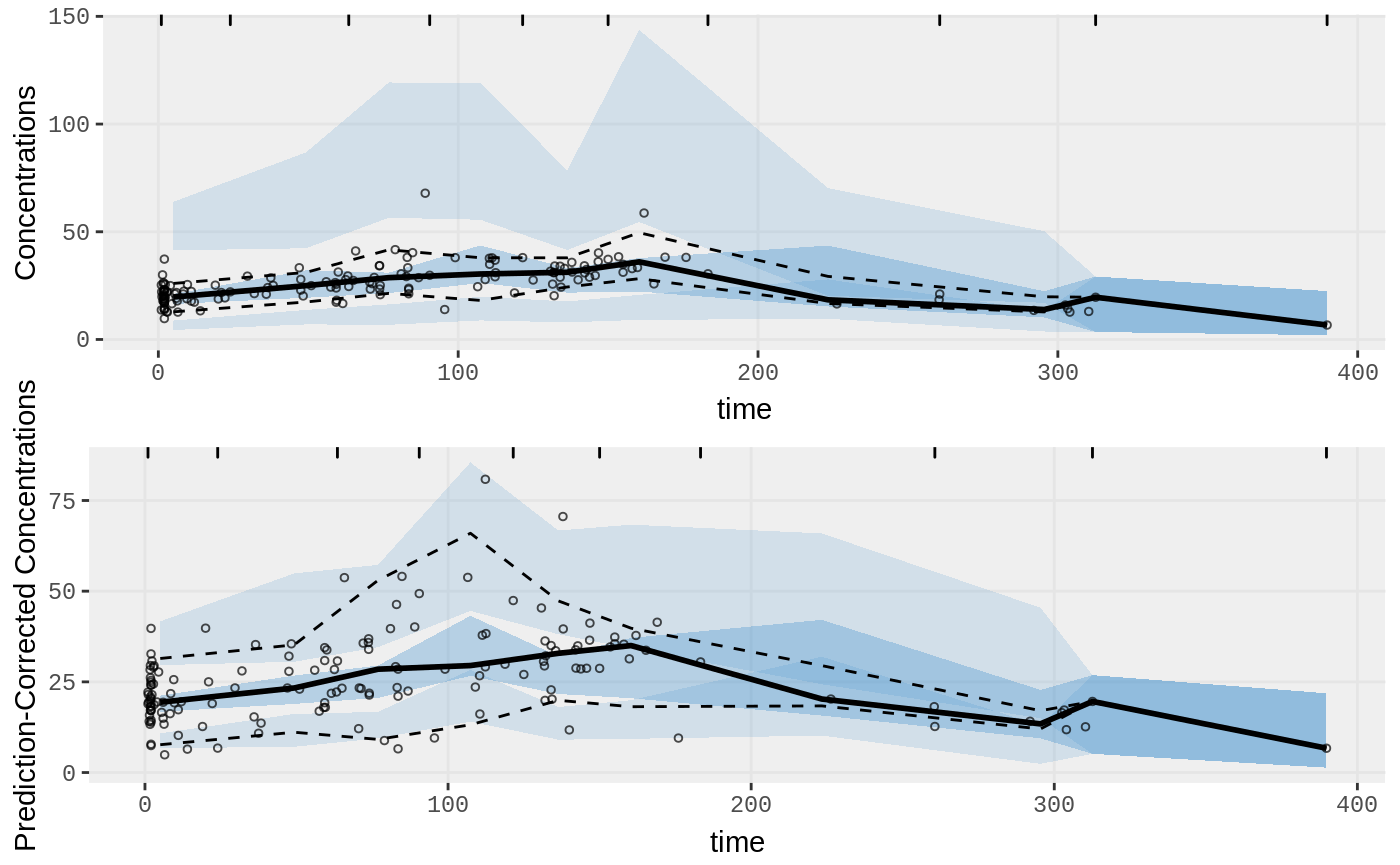
Two types of VPCs
library(ggplot2)
p1 <- nlmixr::vpc(fit, show=list(obs_dv=TRUE));
p1 <- p1+ ylab("Concentrations")
## A prediction-corrected VPC
p2 <- nlmixr::vpc(fit, pred_corr = TRUE, show=list(obs_dv=TRUE))
p2 <- p2+ ylab("Prediction-Corrected Concentrations")
library(gridExtra)
grid.arrange(p1,p2)